Association between missense variants of uncertain significance in the CHEK2 gene and hereditary breast cancer: a cosegregation and bioinformatics analysis
- PMID: 38476463
- PMCID: PMC10927753
- DOI: 10.3389/fgene.2023.1274108
Association between missense variants of uncertain significance in the CHEK2 gene and hereditary breast cancer: a cosegregation and bioinformatics analysis
Abstract
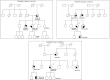
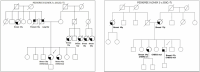
Inherited mutations in the CHEK2 gene have been associated with an increased lifetime risk of developing breast cancer (BC). We aim to identify in the study population the prevalence of mutations in the CHEK2 gene in diagnosed BC patients, evaluate the phenotypic characteristics of the tumor and family history, and predict the deleteriousness of the variants of uncertain significance (VUS). A genetic study was performed, from May 2016 to April 2020, in 396 patients diagnosed with BC at the University Hospital Lozano Blesa of Zaragoza, Spain. Patients with a genetic variant in the CHEK2 gene were selected for the study. We performed a descriptive analysis of the clinical variables, a bibliographic review of the variants, and a cosegregation study when possible. Moreover, an in-depth bioinformatics analysis of CHEK2 VUS was carried out. We identified nine genetic variants in the CHEK2 gene in 10 patients (two pathogenic variants and seven VUS). This supposes a prevalence of 0.75% and 1.77%, respectively. In all cases, there was a family history of BC in first- and/or second-degree relatives. We carried out a cosegregation study in two families, being positive in one of them. The bioinformatics analyses predicted the pathogenicity of six of the VUS. In conclusion, CHEK2 mutations have been associated with an increased risk for BC. This risk is well-established for foundation variants. However, the risk assessment for other variants is unclear. The incorporation of bioinformatics analysis provided supporting evidence of the pathogenicity of VUS.
Keywords: CHEK2; bioinformatics analysis; breast cancer; cancer genetics; genetic testing.
Copyright © 2024 Alonso, Menao, Lastra, Arruebo, Bueso, Pérez, Murillo, Álvarez, Alonso, Rebollar, Cruellas, Arribas, Ramos, Isla, Galano-Frutos, García-Cebollada, Sancho and Andrés.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Balmaña J., Digiovanni L., Gaddam P., Walsh M. F., Joseph V., Stadler Z. K., et al. (2016). Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing. J. Clin. Oncol. 34 (34), 4071–4078. 10.1200/JCO.2016.68.4316 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

