Decoding semiotic minimal genome: a non-genocentric approach
- PMID: 38476952
- PMCID: PMC10929006
- DOI: 10.3389/fmicb.2024.1356050
Decoding semiotic minimal genome: a non-genocentric approach
Abstract
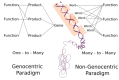
The search for the minimum information required for an organism to sustain a cellular system network has rendered both the identification of a fixed number of known genes and those genes whose function remains to be identified. The approaches used in such search generally focus their analysis on coding genomic regions, based on the genome to proteic-product perspective. Such approaches leave other fundamental processes aside, mainly those that include higher-level information management. To cope with this limitation, a non-genocentric approach based on genomic sequence analysis using language processing tools and gene ontology may prove an effective strategy for the identification of those fundamental genomic elements for life autonomy. Additionally, this approach will provide us with an integrative analysis of the information value present in all genomic elements, regardless of their coding status.
Keywords: biological information flow; genomic semiotics; language processing tools; minimum genome; non-genocentric.
Copyright © 2024 Gómez-Márquez, Morales, Romero-Gutiérrez, Paredes and Borrayo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water-use efficiency in Populus spp.BMC Plant Biol. 2012 Sep 26;12:173. doi: 10.1186/1471-2229-12-173. BMC Plant Biol. 2012. PMID: 23013168 Free PMC article.
-
Primary transcripts: From the discovery of RNA processing to current concepts of gene expression - Review.Exp Cell Res. 2018 Dec 15;373(1-2):1-33. doi: 10.1016/j.yexcr.2018.09.011. Epub 2018 Sep 26. Exp Cell Res. 2018. PMID: 30266658 Review.
-
Genome annotation for clinical genomic diagnostics: strengths and weaknesses.Genome Med. 2017 May 30;9(1):49. doi: 10.1186/s13073-017-0441-1. Genome Med. 2017. PMID: 28558813 Free PMC article. Review.
References
-
- Aguilar-Valdez S., Morales J. A., Paredes O. (2021). “Unraveling the hcov-19 informational architecture,” in 2021 43rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (México: IEEE; ), 2392–2395. 10.1109/EMBC46164.2021.9630954. Available online at: https://ieeexplore.ieee.org/document/9630954 - DOI - PubMed
LinkOut - more resources
Full Text Sources

