Genome sequence diversity of SARS-CoV-2 in Serbia: insights gained from a 3-year pandemic study
- PMID: 38476954
- PMCID: PMC10929721
- DOI: 10.3389/fmicb.2024.1332276
Genome sequence diversity of SARS-CoV-2 in Serbia: insights gained from a 3-year pandemic study
Abstract
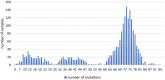
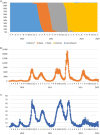
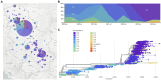
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the COVID-19 pandemic, has been evolving rapidly causing emergence of new variants and health uncertainties. Monitoring the evolution of the virus was of the utmost importance for public health interventions and the development of national and global mitigation strategies. Here, we report national data on the emergence of new variants, their distribution, and dynamics in a 3-year study conducted from March 2020 to the end of January 2023 in the Republic of Serbia. Nasopharyngeal and oropharyngeal swabs from 2,398 COVID-19-positive patients were collected and sequenced using three different next generation technologies: Oxford Nanopore, Ion Torrent, and DNBSeq. In the subset of 2,107 SARS-CoV-2 sequences which met the quality requirements, detection of mutations, assignment to SARS-CoV-2 lineages, and phylogenetic analysis were performed. During the 3-year period, we detected three variants of concern, namely, Alpha (5.6%), Delta (7.4%), and Omicron (70.3%) and one variant of interest-Omicron recombinant "Kraken" (XBB1.5) (<1%), whereas 16.8% of the samples belonged to other SARS-CoV-2 (sub)lineages. The detected SARS-CoV-2 (sub)lineages resulted in eight COVID-19 pandemic waves in Serbia, which correspond to the pandemic waves reported in Europe and the United States. Wave dynamics in Serbia showed the most resemblance with the profile of pandemic waves in southern Europe, consistent with the southeastern European location of Serbia. The samples were assigned to sixteen SARS-CoV-2 Nextstrain clades: 20A, 20B, 20C, 20D, 20E, 20G, 20I, 21J, 21K, 21L, 22A, 22B, 22C, 22D, 22E, and 22F and six different Omicron recombinants (XZ, XAZ, XAS, XBB, XBF, and XBK). The 10 most common mutations detected in the coding and untranslated regions of the SARS-CoV-2 genomes included four mutations affecting the spike protein (S:D614G, S:T478K, S:P681H, and S:S477N) and one mutation at each of the following positions: 5'-untranslated region (5'UTR:241); N protein (N:RG203KR); NSP3 protein (NSP3:F106F); NSP4 protein (NSP4:T492I); NSP6 protein (NSP6: S106/G107/F108 - triple deletion), and NSP12b protein (NSP12b:P314L). This national-level study is the most comprehensive in terms of sequencing and genomic surveillance of SARS-CoV-2 during the pandemic in Serbia, highlighting the importance of establishing and maintaining good national practice for monitoring SARS-CoV-2 and other viruses circulating worldwide.
Keywords: COVID-19; SARS-CoV-2; Serbia; genome; next generation sequencing; pandemic.
Copyright © 2024 Novkovic, Banovic Djeri, Ristivojevic, Knezevic, Jankovic, Tanasic, Radojicic, Keckarevic, Vidanovic, Tesovic, Skakic, Tolinacki, Moric and Djordjevic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
[Genomic Characterization of SARS-CoV-2 Isolates Obtained from Antalya, Türkiye].Mikrobiyol Bul. 2024 Oct;58(4):433-447. doi: 10.5578/mb.20249643. Mikrobiyol Bul. 2024. PMID: 39745215 Turkish.
-
Genetic diversity and genomic epidemiology of SARS-CoV-2 during the first 3 years of the pandemic in Morocco: comprehensive sequence analysis, including the unique lineage B.1.528 in Morocco.Access Microbiol. 2024 Oct 7;6(10):000853.v4. doi: 10.1099/acmi.0.000853.v4. eCollection 2024. Access Microbiol. 2024. PMID: 39376591 Free PMC article.
-
Coronavirus Genomes and Unique Mutations in Structural and Non-Structural Proteins in Pakistani SARS-CoV-2 Delta Variants during the Fourth Wave of the Pandemic.Genes (Basel). 2022 Mar 21;13(3):552. doi: 10.3390/genes13030552. Genes (Basel). 2022. PMID: 35328105 Free PMC article.
-
The effects of amino acid substitution of spike protein and genomic recombination on the evolution of SARS-CoV-2.Front Microbiol. 2023 Jul 25;14:1228128. doi: 10.3389/fmicb.2023.1228128. eCollection 2023. Front Microbiol. 2023. PMID: 37560529 Free PMC article. Review.
Cited by
-
Prognostic Impact of Vaccination, Comorbidity, and Inflammatory Biomarkers on Clinical Outcome in Hospitalized Patients with COVID-19.Biomedicines. 2025 Aug 16;13(8):1995. doi: 10.3390/biomedicines13081995. Biomedicines. 2025. PMID: 40868247 Free PMC article.
-
Vaccine-Induced Humoral and Cellular Response to SARS-CoV-2 in Multiple Sclerosis Patients on Ocrelizumab.Vaccines (Basel). 2025 Apr 30;13(5):488. doi: 10.3390/vaccines13050488. Vaccines (Basel). 2025. PMID: 40432100 Free PMC article.
References
-
- Aksamentov I., Roemer C., Hodcroft E. B., Neher R. A. (2021). Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 6:3773. doi: 10.21105/JOSS.03773 - DOI
-
- APHL . Recommendations for SARS-CoV-2 Sequence Data Quality and Reporting Version 1; (2021). Available at: https://www.aphl.org/programs/preparedness/Crisis-Management/Documents/A... (Accessed August 23, 2023).
-
- Chan J. F., Kok K. H., Zhu Z., Chu H., To K. K., Yuan S., et al. . (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9, 221–236. doi: 10.1080/22221751.2020.1719902, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

