Does regulation hold the key to optimizing lipopeptide production in Pseudomonas for biotechnology?
- PMID: 38476965
- PMCID: PMC10928948
- DOI: 10.3389/fbioe.2024.1363183
Does regulation hold the key to optimizing lipopeptide production in Pseudomonas for biotechnology?
Abstract
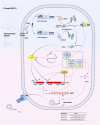
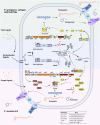
Lipopeptides (LPs) produced by Pseudomonas spp. are specialized metabolites with diverse structures and functions, including powerful biosurfactant and antimicrobial properties. Despite their enormous potential in environmental and industrial biotechnology, low yield and high production cost limit their practical use. While genome mining and functional genomics have identified a multitude of LP biosynthetic gene clusters, the regulatory mechanisms underlying their biosynthesis remain poorly understood. We propose that regulation holds the key to unlocking LP production in Pseudomonas for biotechnology. In this review, we summarize the structure and function of Pseudomonas-derived LPs and describe the molecular basis for their biosynthesis and regulation. We examine the global and specific regulator-driven mechanisms controlling LP synthesis including the influence of environmental signals. Understanding LP regulation is key to modulating production of these valuable compounds, both quantitatively and qualitatively, for industrial and environmental biotechnology.
Keywords: Pseudomonas; antibiotics; bioengineering; bioprocessing; biosurfactants; lipopeptides; regulation; specialized metabolites.
Copyright © 2024 Zhou, Höfte and Hennessy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures







References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

