An anticodon-sensing T-boxzyme generates the elongator nonproteinogenic aminoacyl-tRNA in situ of a custom-made translation system for incorporation
- PMID: 38477328
- PMCID: PMC11039980
- DOI: 10.1093/nar/gkae151
An anticodon-sensing T-boxzyme generates the elongator nonproteinogenic aminoacyl-tRNA in situ of a custom-made translation system for incorporation
Abstract
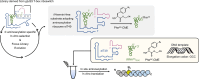
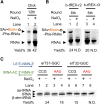
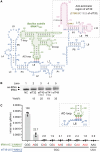
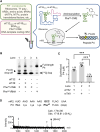
In the hypothetical RNA world, ribozymes could have acted as modern aminoacyl-tRNA synthetases (ARSs) to charge tRNAs, thus giving rise to the peptide synthesis along with the evolution of a primitive translation apparatus. We previously reported a T-boxzyme, Tx2.1, which selectively charges initiator tRNA with N-biotinyl-phenylalanine (BioPhe) in situ in a Flexible In-vitro Translation (FIT) system to produce BioPhe-initiating peptides. Here, we performed in vitro selection of elongation-capable T-boxzymes (elT-boxzymes), using para-azido-l-phenylalanine (PheAZ) as an acyl-donor. We implemented a new strategy to enrich elT-boxzyme-tRNA conjugates that self-aminoacylated on the 3'-terminus selectively. One of them, elT32, can charge PheAZ onto tRNA in trans in response to its cognate anticodon. Further evolution of elT32 resulted in elT49, with enhanced aminoacylation activity. We have demonstrated the translation of a PheAZ-containing peptide in an elT-boxzyme-integrated FIT system, revealing that elT-boxzymes are able to generate the PheAZ-tRNA in response to the cognate anticodon in situ of a custom-made translation system. This study, together with Tx2.1, illustrates a scenario where a series of ribozymes could have overseen aminoacylation and co-evolved with a primitive RNA-based translation system.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
An empirical model of aminoacylation kinetics for E. coli class I and II aminoacyl tRNA synthetases.PLoS Comput Biol. 2025 Aug 12;21(8):e1013353. doi: 10.1371/journal.pcbi.1013353. eCollection 2025 Aug. PLoS Comput Biol. 2025. PMID: 40794817 Free PMC article.
-
Recombinant expression and purification of wheat aminoacyl-tRNA synthetases and IRES-dependent polypeptide synthesis with homogeneously derived purified factors.Biochimie. 2025 Sep;236:30-44. doi: 10.1016/j.biochi.2025.06.010. Epub 2025 Jun 21. Biochimie. 2025. PMID: 40550411
-
An aminoacylation ribozyme evolved from a natural tRNA-sensing T-box riboswitch.Nat Chem Biol. 2020 Jun;16(6):702-709. doi: 10.1038/s41589-020-0500-6. Epub 2020 Mar 23. Nat Chem Biol. 2020. PMID: 32203413
-
Direct and quantitative analysis of tRNA acylation using intact tRNA liquid chromatography-mass spectrometry.Nat Protoc. 2025 May;20(5):1246-1274. doi: 10.1038/s41596-024-01086-9. Epub 2025 Jan 6. Nat Protoc. 2025. PMID: 39762443 Review.
-
Reprogramming the genetic code with flexizymes.Nat Rev Chem. 2024 Dec;8(12):879-892. doi: 10.1038/s41570-024-00656-5. Epub 2024 Oct 21. Nat Rev Chem. 2024. PMID: 39433956 Review.
References
-
- Gilbert W. Origin of life: the RNA world. Nature. 1986; 319:618–618.
-
- Yarus M. A specific amino acid binding site composed of RNA. Science. 1988; 240:1751–1758. - PubMed
-
- Hager A.J., Pollard J.D., Szostak J.W. Ribozymes: aiming at RNA replication and protein synthesis. Chem. Biol. 1996; 3:717–725. - PubMed
-
- Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000; 289:920–930. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

