Gastrointestinal Dysmotility Predisposes to Colitis through Regulation of Gut Microbial Composition and Linoleic Acid Metabolism
- PMID: 38477534
- PMCID: PMC11132037
- DOI: 10.1002/advs.202306297
Gastrointestinal Dysmotility Predisposes to Colitis through Regulation of Gut Microbial Composition and Linoleic Acid Metabolism
Abstract
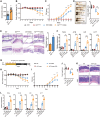
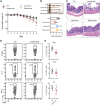
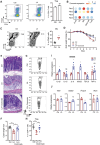
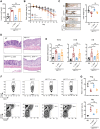
Disrupted gastrointestinal (GI) motility is highly prevalent in patients with inflammatory bowel disease (IBD), but its potential causative role remains unknown. Herein, the role and the mechanism of impaired GI motility in colitis pathogenesis are investigated. Increased colonic mucosal inflammation is found in patients with chronic constipation (CC). Mice with GI dysmotility induced by genetic mutation or chemical insult exhibit increased susceptibility to colitis, dependent on the gut microbiota. GI dysmotility markedly decreases the abundance of Lactobacillus animlalis and increases the abundance of Akkermansia muciniphila. The reduction in L. animlalis, leads to the accumulation of linoleic acid due to compromised conversion to conjugated linoleic acid. The accumulation of linoleic acid inhibits Treg cell differentiation and increases colitis susceptibility via inducing macrophage infiltration and proinflammatory cytokine expression in macrophage. Lactobacillus and A. muciniphila abnormalities are also observed in CC and IBD patients, and mice receiving fecal microbiota from CC patients displayed an increased susceptibility to colitis. These findings suggest that GI dysmotility predisposes host to colitis development by modulating the composition of microbiota and facilitating linoleic acid accumulation. Targeted modulation of microbiota and linoleic acid metabolism may be promising to protect patients with motility disorder from intestinal inflammation.
Keywords: gut microbiota; gut motility; immune cell; inflammatory bowel disease; linoleic acid.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Alatab S., Sepanlou S. G., Ikuta K., Vahedi H., Bisignano C., Safiri S., Sadeghi A., Nixon M. R., Abdoli A., Abolhassani H., Alipour V., Almadi M. A. H., Almasi‐Hashiani A., Anushiravani A., Arabloo J., Atique S., Awasthi A., Badawi A., Baig A. A. A., Bhala N., Bijani A., Biondi A., Borzì A. M., Burke K. E., Carvalho F., Daryani A., Dubey M., Eftekhari A., Fernandes E., Fernandes J. C., et al., The Lancet Gastroenterology & Hepatology 2020, 5, 17. - PubMed
-
- a) Pastor A. C., Osman F., Teitelbaum D. H., Caty M. G., Langer J. C., J. Pediatr. Surg. 2009, 44, 251; - PubMed
- b) Amiel J., Sproat‐Emison E., Garcia‐Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., Brooks A. S., Antinolo G., de Pontual L., Clement‐Ziza M., Munnich A., Kashuk C., West K., Wong K. K.‐Y., Lyonnet S., Chakravarti A., Tam P. K.‐H., Ceccherini I., Hofstra R. M. W., Fernandez R., Journal of Medical Genetics 2008, 45, 1. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
