Hmgcs2 is the hub gene in diabetic cardiomyopathy and is negatively regulated by Hmgcs2, promoting high glucose-induced cardiomyocyte injury
- PMID: 38477658
- PMCID: PMC10936232
- DOI: 10.1002/iid3.1191
Hmgcs2 is the hub gene in diabetic cardiomyopathy and is negatively regulated by Hmgcs2, promoting high glucose-induced cardiomyocyte injury
Abstract
Background: Diabetic cardiomyopathy (DCM) represents a major cause of heart failure and a large medical burden worldwide. This study screened the potentially regulatory targets of DCM and analyzed their roles in high glucose (HG)-induced cardiomyocyte injury.
Methods: Through GEO database, we obtained rat DCM expression chips and screened differentially expressed genes. Rat cardiomyocytes (H9C2) were induced with HG. 3-hydroxy-3-methylglutarylcoenzyme A synthase 2 (Hmgcs2) and microRNA (miR)-363-5p expression patterns in cells were measured by real-time quantitative polymerase chain reaction or Western blot assay, with the dual-luciferase assay to analyze their binding relationship. Then, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide assay, lactate dehydrogenase assay, terminal deoxynucleotidyl transferase dUTP nick end labeling assay, enzyme-linked immunosorbent assay, and various assay kits were applied to evaluate cell viability, cytotoxicity, apoptosis, inflammation responses, and oxidative burden.
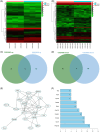
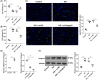
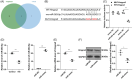
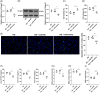
Results: Hmgcs2 was the vital hub gene in DCM. Hmgcs2 was upregulated in HG-induced cardiomyocytes. Hmgcs2 downregulation increased cell viability, decreased TUNEL-positive cell number, reduced HG-induced inflammation and oxidative stress. miR-363-5p is the upstream miRNA of Hmgcs2. miR-363-5p overexpression attenuated HG-induced cell injury.
Conclusions: Hmgcs2 had the most critical regulatory role in DCM. We for the first time reported that miR-363-5p inhibited Hmgcs2 expression, thereby alleviating HG-induced cardiomyocyte injury.
Keywords: GEO database chip; diabetic cardiomyopathy; high glucose; hmgcs2; miR-363-5p.
© 2024 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare that there is no conflict of interests in this study.
Figures

Similar articles
-
HMGCS2 silencing attenuates high glucose-induced in vitro diabetic cardiomyopathy by increasing cell viability, and inhibiting apoptosis, inflammation, and oxidative stress.Bioengineered. 2022 May;13(5):11417-11429. doi: 10.1080/21655979.2022.2063222. Bioengineered. 2022. PMID: 35506308 Free PMC article.
-
Silencing of peroxisome proliferator-activated receptor-alpha alleviates myocardial injury in diabetic cardiomyopathy by downregulating 3-hydroxy-3-methylglutaryl-coenzyme A synthase 2 expression.IUBMB Life. 2020 Sep;72(9):1997-2009. doi: 10.1002/iub.2337. Epub 2020 Jul 30. IUBMB Life. 2020. PMID: 32734614
-
Long Noncoding RNA OIP5-AS1 Overexpression Promotes Viability and Inhibits High Glucose-Induced Oxidative Stress of Cardiomyocytes by Targeting MicroRNA-34a/SIRT1 Axis in Diabetic Cardiomyopathy.Endocr Metab Immune Disord Drug Targets. 2021;21(11):2017-2027. doi: 10.2174/1871530321666201230090742. Endocr Metab Immune Disord Drug Targets. 2021. PMID: 33380310
-
Therapeutic overexpression of miR-92a-2-5p ameliorated cardiomyocyte oxidative stress injury in the development of diabetic cardiomyopathy.Cell Mol Biol Lett. 2022 Oct 8;27(1):85. doi: 10.1186/s11658-022-00379-9. Cell Mol Biol Lett. 2022. PMID: 36209049 Free PMC article.
-
Protein PDK4 Interacts with HMGCS2 to Facilitate High Glucoseinduced Myocardial Injuries.Curr Mol Med. 2023;23(10):1104-1115. doi: 10.2174/1566524023666221021124202. Curr Mol Med. 2023. PMID: 36281857
Cited by
-
JAK2/STAT3/HMGCS2 signaling aggravates mitochondrial dysfunction and oxidative stress in hyperuricemia-induced cardiac dysfunction.Mol Med. 2025 May 13;31(1):184. doi: 10.1186/s10020-025-01246-x. Mol Med. 2025. PMID: 40361044 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- SDkeylab-Endo & LiMe2019-01/Shandong Provincial Key Laboratory of Endocrinology and Lipid Metabolism
- 2019QL017/Medical as well as Academic Promotion Program of Shandong First Medical University
- ZR2017LH023/Natural Science Foundation of Shandong Province of China
- J17KA246/Higher Educational Science and Technology Program of Shandong Province, China
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

