Neuronal STING activation in amyotrophic lateral sclerosis and frontotemporal dementia
- PMID: 38478117
- PMCID: PMC10937762
- DOI: 10.1007/s00401-024-02688-z
Neuronal STING activation in amyotrophic lateral sclerosis and frontotemporal dementia
Abstract
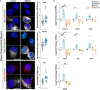
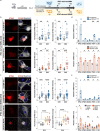
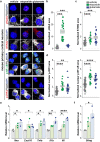
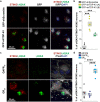
The stimulator of interferon genes (STING) pathway has been implicated in neurodegenerative diseases, including Parkinson's disease and amyotrophic lateral sclerosis (ALS). While prior studies have focused on STING within immune cells, little is known about STING within neurons. Here, we document neuronal activation of the STING pathway in human postmortem cortical and spinal motor neurons from individuals affected by familial or sporadic ALS. This process takes place selectively in the most vulnerable cortical and spinal motor neurons but not in neurons that are less affected by the disease. Concordant STING activation in layer V cortical motor neurons occurs in a mouse model of C9orf72 repeat-associated ALS and frontotemporal dementia (FTD). To establish that STING activation occurs in a neuron-autonomous manner, we demonstrate the integrity of the STING signaling pathway, including both upstream activators and downstream innate immune response effectors, in dissociated mouse cortical neurons and neurons derived from control human induced pluripotent stem cells (iPSCs). Human iPSC-derived neurons harboring different familial ALS-causing mutations exhibit increased STING signaling with DNA damage as a main driver. The elevated downstream inflammatory markers present in ALS iPSC-derived neurons can be suppressed with a STING inhibitor. Our results reveal an immunophenotype that consists of innate immune signaling driven by the STING pathway and occurs specifically within vulnerable neurons in ALS/FTD.
Keywords: Amyotrophic lateral sclerosis; Central neurons; DNA damage; Motor neuron disease; Neuroinflammation; STING.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests. B.T.H. has a family member who works at Novartis and owns stock in Novartis; he serves on the SAB of Dewpoint and owns stock. He serves on a scientific advisory board or is a consultant for AbbVie, Aprinoia Therapeutics, Arvinas, Avrobio, Axial, Biogen, BMS, Cure Alz Fund, Cell Signaling, Eisai, Genentech, Ionis, Latus, Novartis, Sangamo, Sanofi, Seer, Takeda, the US Dept. of Justice, Vigil, and Voyager. His laboratory is supported by research grants from the National Institutes of Health, Cure Alzheimer’s Fund, Tau Consortium, and the JPB Foundation and sponsored research agreements from Abbvie, BMS, and Biogen. L.P. and Mayo Clinic have licensed technology involving
Figures



References
-
- Abo-Rady M, Kalmbach N, Pal A, Schludi C, Janosch A, Richter T, Freitag P, Bickle M, Kahlert AK, Petri S, et al. Knocking out C9ORF72 Exacerbates Axonal Trafficking Defects Associated with Hexanucleotide Repeat Expansion and Reduces Levels of Heat Shock Proteins. Stem Cell Reports. 2020;14:390–405. doi: 10.1016/j.stemcr.2020.01.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

