Coordination of histone chaperones for parental histone segregation and epigenetic inheritance
- PMID: 38479839
- PMCID: PMC10982699
- DOI: 10.1101/gad.351278.123
Coordination of histone chaperones for parental histone segregation and epigenetic inheritance
Abstract
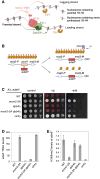
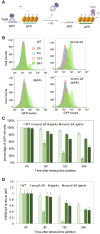
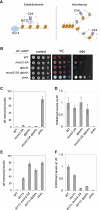
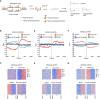
Chromatin-based epigenetic memory relies on the accurate distribution of parental histone H3-H4 tetramers to newly replicated DNA strands. Mcm2, a subunit of the replicative helicase, and Dpb3/4, subunits of DNA polymerase ε, govern parental histone H3-H4 deposition to the lagging and leading strands, respectively. However, their contribution to epigenetic inheritance remains controversial. Here, using fission yeast heterochromatin inheritance systems that eliminate interference from initiation pathways, we show that a Mcm2 histone binding mutation severely disrupts heterochromatin inheritance, while mutations in Dpb3/4 cause only moderate defects. Surprisingly, simultaneous mutations of Mcm2 and Dpb3/4 stabilize heterochromatin inheritance. eSPAN (enrichment and sequencing of protein-associated nascent DNA) analyses confirmed the conservation of Mcm2 and Dpb3/4 functions in parental histone H3-H4 segregation, with their combined absence showing a more symmetric distribution of parental histone H3-H4 than either single mutation alone. Furthermore, the FACT histone chaperone regulates parental histone transfer to both strands and collaborates with Mcm2 and Dpb3/4 to maintain parental histone H3-H4 density and faithful heterochromatin inheritance. These results underscore the importance of both symmetric distribution of parental histones and their density at daughter strands for epigenetic inheritance and unveil distinctive properties of parental histone chaperones during DNA replication.
Keywords: Dpb3; Dpb4; H3K9 methylation; Mcm2; eSPAN; epigenetic inheritance; fission yeast; heterochromatin; histone chaperone; parental histone density.
© 2024 Fang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance.Biology (Basel). 2024 Jul 29;13(8):572. doi: 10.3390/biology13080572. Biology (Basel). 2024. PMID: 39194510 Free PMC article. Review.
-
Mrc1 regulates parental histone segregation and heterochromatin inheritance.Mol Cell. 2024 Sep 5;84(17):3223-3236.e4. doi: 10.1016/j.molcel.2024.07.002. Epub 2024 Aug 1. Mol Cell. 2024. PMID: 39094566
-
Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):12524-12529. doi: 10.1073/pnas.1712961114. Epub 2017 Nov 6. Proc Natl Acad Sci U S A. 2017. PMID: 29109278 Free PMC article.
-
A mechanism for preventing asymmetric histone segregation onto replicating DNA strands.Science. 2018 Sep 28;361(6409):1386-1389. doi: 10.1126/science.aat8849. Epub 2018 Aug 16. Science. 2018. PMID: 30115745 Free PMC article.
-
How is epigenetic information on chromatin inherited after DNA replication?Ernst Schering Res Found Workshop. 2006;(57):89-96. doi: 10.1007/3-540-37633-x_5. Ernst Schering Res Found Workshop. 2006. PMID: 16568950 Review.
Cited by
-
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability.Sci Adv. 2025 Feb 21;11(8):eadr1453. doi: 10.1126/sciadv.adr1453. Epub 2025 Feb 19. Sci Adv. 2025. PMID: 39970210 Free PMC article.
-
The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance.Biology (Basel). 2024 Jul 29;13(8):572. doi: 10.3390/biology13080572. Biology (Basel). 2024. PMID: 39194510 Free PMC article. Review.
-
A tale of two strands: Decoding chromatin replication through strand-specific sequencing.Mol Cell. 2025 Jan 16;85(2):238-261. doi: 10.1016/j.molcel.2024.10.035. Epub 2025 Jan 16. Mol Cell. 2025. PMID: 39824166 Review.
-
Mrc1 regulates parental histone segregation and heterochromatin inheritance.Mol Cell. 2024 Sep 5;84(17):3223-3236.e4. doi: 10.1016/j.molcel.2024.07.002. Epub 2024 Aug 1. Mol Cell. 2024. PMID: 39094566
-
Insights into the synchronization between DNA replication and parental histone recycling.Biochem Soc Trans. 2025 Jun 30;53(3):547-554. doi: 10.1042/BST20253014. Biochem Soc Trans. 2025. PMID: 40380884 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
