Charting cellular differentiation trajectories with Ricci flow
- PMID: 38480714
- PMCID: PMC10937996
- DOI: 10.1038/s41467-024-45889-6
Charting cellular differentiation trajectories with Ricci flow
Abstract
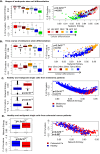
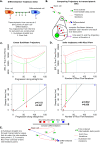
Complex biological processes, such as cellular differentiation, require intricate rewiring of intra-cellular signalling networks. Previous characterisations revealed a raised network entropy underlies less differentiated and malignant cell states. A connection between entropy and Ricci curvature led to applications of discrete curvatures to biological networks. However, predicting dynamic biological network rewiring remains an open problem. Here we apply Ricci curvature and Ricci flow to biological network rewiring. By investigating the relationship between network entropy and Forman-Ricci curvature, theoretically and empirically on single-cell RNA-sequencing data, we demonstrate that the two measures do not always positively correlate, as previously suggested, and provide complementary rather than interchangeable information. We next employ Ricci flow to derive network rewiring trajectories from stem cells to differentiated cells, accurately predicting true intermediate time points in gene expression time courses. In summary, we present a differential geometry toolkit for understanding dynamic network rewiring during cellular differentiation and cancer.
© 2024. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Discrete Ricci curvatures capture age-related changes in human brain functional connectivity networks.Front Aging Neurosci. 2023 May 24;15:1120846. doi: 10.3389/fnagi.2023.1120846. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37293668 Free PMC article.
-
Comparative analysis of two discretizations of Ricci curvature for complex networks.Sci Rep. 2018 Jun 5;8(1):8650. doi: 10.1038/s41598-018-27001-3. Sci Rep. 2018. PMID: 29872167 Free PMC article.
-
Network Geometry of Borsa Istanbul: Analyzing Sectoral Dynamics with Forman-Ricci Curvature.Entropy (Basel). 2025 Mar 5;27(3):271. doi: 10.3390/e27030271. Entropy (Basel). 2025. PMID: 40149195 Free PMC article.
-
Energy management - a critical role in cancer induction?Crit Rev Oncol Hematol. 2013 Oct;88(1):198-217. doi: 10.1016/j.critrevonc.2013.04.001. Epub 2013 May 31. Crit Rev Oncol Hematol. 2013. PMID: 23731619 Review.
-
Epigenome modifiers and metabolic rewiring: New frontiers in therapeutics.Pharmacol Ther. 2019 Jan;193:178-193. doi: 10.1016/j.pharmthera.2018.08.008. Epub 2018 Aug 17. Pharmacol Ther. 2019. PMID: 30125527 Review.
Cited by
-
Deep learning as Ricci flow.Sci Rep. 2024 Oct 8;14(1):23383. doi: 10.1038/s41598-024-74045-9. Sci Rep. 2024. PMID: 39379488 Free PMC article.
-
Cellular Development Follows the Path of Minimum Action.ArXiv [Preprint]. 2025 Apr 10:arXiv:2504.08096v1. ArXiv. 2025. PMID: 40297223 Free PMC article. Preprint.
-
The network structural entropy for single-cell RNA sequencing data during skin aging.Brief Bioinform. 2024 Nov 22;26(1):bbae698. doi: 10.1093/bib/bbae698. Brief Bioinform. 2024. PMID: 39757115 Free PMC article.
References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
-
- Waddington, C. H. An Introduction to Modern Genetics. (George Alien & Unwin, London,1939)
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

