Natural variation in OsMYB8 confers diurnal floret opening time divergence between indica and japonica subspecies
- PMID: 38480732
- PMCID: PMC10937712
- DOI: 10.1038/s41467-024-46579-z
Natural variation in OsMYB8 confers diurnal floret opening time divergence between indica and japonica subspecies
Abstract
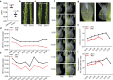
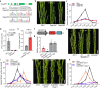
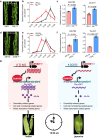
The inter-subspecific indica-japonica hybrid rice confer potential higher yield than the widely used indica-indica intra-subspecific hybrid rice. Nevertheless, the utilization of this strong heterosis is currently hindered by asynchronous diurnal floret opening time (DFOT) of indica and japonica parental lines. Here, we identify OsMYB8 as a key regulator of rice DFOT. OsMYB8 induces the transcription of JA-Ile synthetase OsJAR1, thereby regulating the expression of genes related to cell osmolality and cell wall remodeling in lodicules to promote floret opening. Natural variations of OsMYB8 promoter contribute to its differential expression, thus differential transcription of OsJAR1 and accumulation of JA-Ile in lodicules of indica and japonica subspecies. Furthermore, introgression of the indica haplotype of OsMYB8 into japonica effectively promotes DFOT in japonica. Our findings reveal an OsMYB8-OsJAR1 module that regulates differential DFOT in indica and japonica, and provide a strategy for breeding early DFOT japonica to facilitate breeding of indica-japonica hybrids.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Ma G, Yuan L. Hybrid rice achievements, development and prospect in China. J. Integr. Agr. 2015;14:197–205. doi: 10.1016/S2095-3119(14)60922-9. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

