Dendritic cell-targeted therapy expands CD8 T cell responses to bona-fide neoantigens in lung tumors
- PMID: 38480738
- PMCID: PMC10937682
- DOI: 10.1038/s41467-024-46685-y
Dendritic cell-targeted therapy expands CD8 T cell responses to bona-fide neoantigens in lung tumors
Abstract
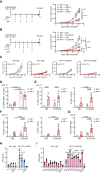
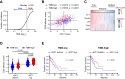
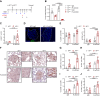
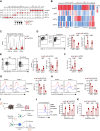
Cross-presentation by type 1 DCs (cDC1) is critical to induce and sustain antitumoral CD8 T cell responses to model antigens, in various tumor settings. However, the impact of cross-presenting cDC1 and the potential of DC-based therapies in tumors carrying varied levels of bona-fide neoantigens (neoAgs) remain unclear. Here we develop a hypermutated model of non-small cell lung cancer in female mice, encoding genuine MHC-I neoepitopes to study neoAgs-specific CD8 T cell responses in spontaneous settings and upon Flt3L + αCD40 (DC-therapy). We find that cDC1 are required to generate broad CD8 responses against a range of diverse neoAgs. DC-therapy promotes immunogenicity of weaker neoAgs and strongly inhibits the growth of high tumor-mutational burden (TMB) tumors. In contrast, low TMB tumors respond poorly to DC-therapy, generating mild CD8 T cell responses that are not sufficient to block progression. scRNA transcriptional analysis, immune profiling and functional assays unveil the changes induced by DC-therapy in lung tissues, which comprise accumulation of cDC1 with increased immunostimulatory properties and less exhausted effector CD8 T cells. We conclude that boosting cDC1 activity is critical to broaden the diversity of anti-tumoral CD8 T cell responses and to leverage neoAgs content for therapeutic advantage.
© 2024. The Author(s).
Conflict of interest statement
A.B. and G.G. are cofounders and shareholders of NeoPhore. A.B. is a member of the NeoPhore scientific advisory board. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

