Functional genomic screens with death rate analyses reveal mechanisms of drug action
- PMID: 38480981
- PMCID: PMC11393183
- DOI: 10.1038/s41589-024-01584-7
Functional genomic screens with death rate analyses reveal mechanisms of drug action
Abstract
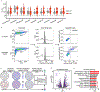
A common approach for understanding how drugs induce their therapeutic effects is to identify the genetic determinants of drug sensitivity. Because 'chemo-genetic profiles' are performed in a pooled format, inference of gene function is subject to several confounding influences related to variation in growth rates between clones. In this study, we developed Method for Evaluating Death Using a Simulation-assisted Approach (MEDUSA), which uses time-resolved measurements, along with model-driven constraints, to reveal the combination of growth and death rates that generated the observed drug response. MEDUSA is uniquely effective at identifying death regulatory genes. We apply MEDUSA to characterize DNA damage-induced lethality in the presence and absence of p53. Loss of p53 switches the mechanism of DNA damage-induced death from apoptosis to a non-apoptotic death that requires high respiration. These findings demonstrate the utility of MEDUSA both for determining the genetic dependencies of lethality and for revealing opportunities to potentiate chemo-efficacy in a cancer-specific manner.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures








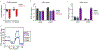

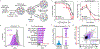




References
METHODS REFERENCES
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

