Centralized Interactive Phenomics Resource: an integrated online phenomics knowledgebase for health data users
- PMID: 38481028
- PMCID: PMC11031216
- DOI: 10.1093/jamia/ocae042
Centralized Interactive Phenomics Resource: an integrated online phenomics knowledgebase for health data users
Abstract
Objective: Development of clinical phenotypes from electronic health records (EHRs) can be resource intensive. Several phenotype libraries have been created to facilitate reuse of definitions. However, these platforms vary in target audience and utility. We describe the development of the Centralized Interactive Phenomics Resource (CIPHER) knowledgebase, a comprehensive public-facing phenotype library, which aims to facilitate clinical and health services research.
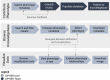
Materials and methods: The platform was designed to collect and catalog EHR-based computable phenotype algorithms from any healthcare system, scale metadata management, facilitate phenotype discovery, and allow for integration of tools and user workflows. Phenomics experts were engaged in the development and testing of the site.
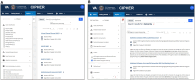
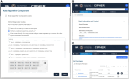
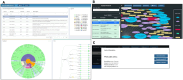
Results: The knowledgebase stores phenotype metadata using the CIPHER standard, and definitions are accessible through complex searching. Phenotypes are contributed to the knowledgebase via webform, allowing metadata validation. Data visualization tools linking to the knowledgebase enhance user interaction with content and accelerate phenotype development.
Discussion: The CIPHER knowledgebase was developed in the largest healthcare system in the United States and piloted with external partners. The design of the CIPHER website supports a variety of front-end tools and features to facilitate phenotype development and reuse. Health data users are encouraged to contribute their algorithms to the knowledgebase for wider dissemination to the research community, and to use the platform as a springboard for phenotyping.
Conclusion: CIPHER is a public resource for all health data users available at https://phenomics.va.ornl.gov/ which facilitates phenotype reuse, development, and dissemination of phenotyping knowledge.
Keywords: algorithms; electronic health records; library; phenomics.
Published by Oxford University Press on behalf of the American Medical Informatics Association 2024.
Conflict of interest statement
The authors have no competing interests to declare.
Figures

References
-
- Huang H, Turner M, Raju S, et al.Identification of acute decompensated heart failure hospitalizations using administrative data. Am J Cardiol. 2017;119(11):1791-1796. - PubMed

