Whole genome sequencing increases the diagnostic rate in Charcot-Marie-Tooth disease
- PMID: 38481354
- PMCID: PMC11370804
- DOI: 10.1093/brain/awae064
Whole genome sequencing increases the diagnostic rate in Charcot-Marie-Tooth disease
Abstract
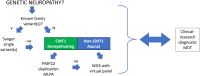
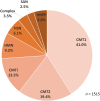
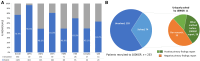
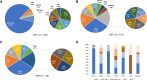
Charcot-Marie-Tooth disease (CMT) is one of the most common and genetically heterogeneous inherited neurological diseases, with more than 130 disease-causing genes. Whole genome sequencing (WGS) has improved diagnosis across genetic diseases, but the diagnostic impact in CMT is yet to be fully reported. We present the diagnostic results from a single specialist inherited neuropathy centre, including the impact of WGS diagnostic testing. Patients were assessed at our specialist inherited neuropathy centre from 2009 to 2023. Genetic testing was performed using single gene testing, next-generation sequencing targeted panels, research whole exome sequencing and WGS and, latterly, WGS through the UK National Health Service. Variants were assessed using the American College of Medical Genetics and Genomics and Association for Clinical Genomic Science criteria. Excluding patients with hereditary ATTR amyloidosis, 1515 patients with a clinical diagnosis of CMT and related disorders were recruited. In summary, 621 patients had CMT1 (41.0%), 294 CMT2 (19.4%), 205 intermediate CMT (CMTi, 13.5%), 139 hereditary motor neuropathy (HMN, 9.2%), 93 hereditary sensory neuropathy (HSN, 6.1%), 38 sensory ataxic neuropathy (2.5%), 72 hereditary neuropathy with liability to pressure palsies (HNPP, 4.8%) and 53 'complex' neuropathy (3.5%). Overall, a genetic diagnosis was reached in 76.9% (1165/1515). A diagnosis was most likely in CMT1 (96.8%, 601/621), followed by CMTi (81.0%, 166/205) and then HSN (69.9%, 65/93). Diagnostic rates remained less than 50% in CMT2, HMN and complex neuropathies. The most common genetic diagnosis was PMP22 duplication (CMT1A; 505/1165, 43.3%), then GJB1 (CMTX1; 151/1165, 13.0%), PMP22 deletion (HNPP; 72/1165, 6.2%) and MFN2 (CMT2A; 46/1165, 3.9%). We recruited 233 cases to the UK 100 000 Genomes Project (100KGP), of which 74 (31.8%) achieved a diagnosis; 28 had been otherwise diagnosed since recruitment, leaving a true diagnostic rate of WGS through the 100KGP of 19.7% (46/233). However, almost half of the solved cases (35/74) received a negative report from the study, and the diagnosis was made through our research access to the WGS data. The overall diagnostic uplift of WGS for the entire cohort was 3.5%. Our diagnostic rate is the highest reported from a single centre and has benefitted from the use of WGS, particularly access to the raw data. However, almost one-quarter of all cases remain unsolved, and a new reference genome and novel technologies will be important to narrow the 'diagnostic gap'.
Keywords: CMT; HMSN; WES; WGS; genomics; neurogenetics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Reilly MM. Sorting out the inherited neuropathies. Pract Neurol. 2007;7:93–105. - PubMed
-
- Laurá M, Pipis M, Rossor AM, Reilly MM. Charcot-Marie-Tooth disease and related disorders: an evolving landscape. Curr Opin Neurol. 2019;32:641–650. - PubMed
-
- Rossor AM, Carr AS, Devine H, et al. Peripheral neuropathy in complex inherited diseases: an approach to diagnosis. J Neurol Neurosurg Psychiatry. 2017;88:846–863. - PubMed
-
- Masingue M, Fernández-Eulate G, Debs R, et al. Strategy for genetic analysis in hereditary neuropathy. Rev Neurol (Paris). 2022;9:10–29. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

