Metabolic engineering of Streptomyces peucetius for biosynthesis of N,N-dimethylated anthracyclines
- PMID: 38481571
- PMCID: PMC10936713
- DOI: 10.3389/fbioe.2024.1363803
Metabolic engineering of Streptomyces peucetius for biosynthesis of N,N-dimethylated anthracyclines
Abstract
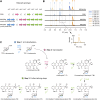
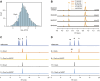
Introduction: Daunorubicin and doxorubicin, two anthracycline polyketides produced by S. peucetius, are potent anticancer agents that are widely used in chemotherapy, despite severe side effects. Recent advances have highlighted the potential of producing improved derivatives with reduced side effects by incorporating l-rhodosamine, the N,N-dimethyl analogue of the native amino sugar moiety. Method: In this study, we aimed to produce N,N-dimethylated anthracyclines by engineering the doxorubicin biosynthetic pathway in the industrial Streptomyces peucetius strain G001. To achieve this, we introduced genes from the aclarubicin biosynthetic pathway encoding the sugar N-methyltransferases AclP and AknX2. Furthermore, the native gene for glycosyltransferase DnrS was replaced with genes encoding the aclarubicin glycosyltransferases AknS and AknT. Additionally, the gene for methylesterase RdmC from the rhodomycin biosynthetic pathway was introduced. Results: A new host was engineered successfully, whereby genes from the aclarubicin pathway were introduced and expressed. LC-MS/MS analysis of the engineered strains showed that dimethylated sugars were efficiently produced, and that these were incorporated ino the anthracycline biosynthetic pathway to produce the novel dimethylated anthracycline N,N-dimethyldaunorubicin. Further downstream tailoring steps catalysed by the cytochrome P450 monooxygenase DoxA exhibited limited efficacy with N,N-dimethylated substrates. This resulted in only low production levels of N,N-dimethyldaunorubicin and no N,N-dimethyldoxorubicin, most likely due to the low affinity of DoxA for dimethylated substrates. Discussion: S. peucetius G001 was engineered such as to produce N,N-dimethylated sugars, which were incorporated into the biosynthetic pathway. This allowed the successful production of N,N-dimethyldaunorubicin, an anticancer drug with reduced cytotoxicity. DoxA is the key enzyme that determines the efficiency of the biosynthesis of N,N-dimethylated anthracyclines, and engineering of this enzyme will be a major step forwards towards the efficient production of more N,N-dimethylated anthracyclines, including N,N-dimethyldoxorubicin. This study provides valuable insights into the biosynthesis of clinically relevant daunorubicin derivatives, highlighting the importance of combinatorial biosynthesis.
Keywords: Streptomyces; anthracyclines; anticancer; biosynthesis; doxorubicin; metabolic engineering.
Copyright © 2024 Hulst, Zhang, van der Heul, Du, Elsayed, Koroleva, Grocholski, Wander, Metsä-Ketelä, Neefjes and van Wezel.
Conflict of interest statement
JN is a shareholder in NIHM that aims to produce aclarubicin for clinical use. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

