Defining a metagenomic threshold for detecting low abundances of Providencia alcalifaciens in canine faecal samples
- PMID: 38481663
- PMCID: PMC10933104
- DOI: 10.3389/fcimb.2024.1305742
Defining a metagenomic threshold for detecting low abundances of Providencia alcalifaciens in canine faecal samples
Abstract
Introduction: Acute haemorrhagic diarrhoea syndrome (AHDS) in dogs is a condition of unknown aetiology. Providencia alcalifaciens is suspected to play a role in the disease as it was commonly found in dogs suffering from AHDS during a Norwegian outbreak in 2019. The role of this bacterium as a constituent of the canine gut microbiota is unknown, hence this study set out to investigate its occurrence in healthy dogs using metagenomics.
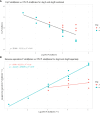
Materials and methods: To decrease the likelihood of false detection, we established a metagenomic threshold for P. alcalifaciens by spiking culture-negative stool samples with a range of bacterial dilutions and analysing these by qPCR and shotgun metagenomics. The detection limit for P. alcalifaciens was determined and used to establish a metagenomic threshold. The threshold was validated on naturally contaminated faecal samples with known cultivation status for P. alcalifaciens. Finally, the metagenomic threshold was used to determine the occurrence of P. alcalifaciens in shotgun metagenomic datasets from canine faecal samples (n=362) collected in the HUNT One Health project.
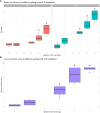
Results: The metagenomic assay and qPCR had a detection limit of 1.1x103 CFU P. alcalifaciens per faecal sample, which corresponded to a Cq value of 31.4 and 569 unique k-mer counts by shotgun metagenomics. Applying this metagenomic threshold to 362 faecal metagenomic datasets from healthy dogs, P. alcalifaciens was found in only 1.1% (95% CI [0.0, 6.8]) of the samples, and then in low relative abundances (median: 0.04%; range: 0.00 to 0.81%). The sensitivity of the qPCR and shotgun metagenomics assay was low, as only 40% of culture-positive samples were also positive by qPCR and metagenomics.
Discussion: Using our detection limit, the occurrence of P. alcalifaciens in faecal samples from healthy dogs was low. Given the low sensitivity of the metagenomic assay, these results do not rule out a significantly higher occurrence of this bacterium at a lower abundance.
Keywords: AHDS; canine; clinical metagenomics; detection limit; faecal microbiota; shotgun sequencing.
Copyright © 2024 Aardal, Soltvedt, Nørstebø, Haverkamp, Rodriguez-Campos, Skancke and Llarena.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adeolu M., Alnajar S., Naushad S., S. Gupta R. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66 (12), 5575–5599. doi: 10.1099/ijsem.0.001485 - DOI - PubMed
-
- Andrews S. (2023). FastQC. Available at: https://github.com/s-andrews/FastQC.
-
- Aphalo P. (2023). ggpmisc: Miscellaneous Extensions to ‘ggplot2’. Available at: https://docs.r4photobiology.info/ggpmisc/, https://github.com/aphalo/ggpmisc.
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

