Identification of transport systems involved in eflornithine delivery across the blood-brain barrier
- PMID: 38482132
- PMCID: PMC7615738
- DOI: 10.3389/fddev.2023.1113493
Identification of transport systems involved in eflornithine delivery across the blood-brain barrier
Abstract
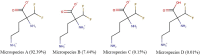
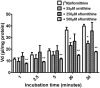
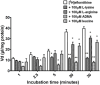
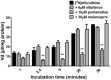
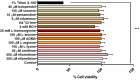
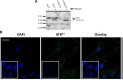
Human African Trypanosomiasis (HAT) is a neglected parasitic disease that continues to persist in sub-Saharan Africa. It is fatal if untreated. The first stage of the disease is associated with the presence of the parasite in the periphery and the second stage with the presence of the parasites in the CNS. The treatment of CNS stage HAT requires the drugs to cross the blood-brain barrier (BBB). Eflornithine is an amino acid analogue that is used to treat second stage HAT gambiense both alone and in combination with nifurtimox. Recent studies have identified that accumulation of eflornithine into the parasites (trypanosomes) involves the amino acid transporter (Trypanosoma brucei AAT6). In this study we tested the hypothesis that eflornithine uses a cationic amino acid transport system to cross the BBB. We particularly focused on system-y+ and system-B0,+. To do this we utilized specialist databases to compare the physicochemical characteristics of relevant molecules and an in vitro model of the BBB to explore the mechanisms of eflornithine delivery into the CNS. Our results confirmed that eflornithine is related to the endogenous amino acid, ornithine. At pH 7.4, eflornithine is predominately (92.39%) a zwitterionic (dipolar) amino acid and ornithine is predominately (99.08%) a cationic (tripolar) amino acid. In addition, the gross charge distribution at pH 7.4 of eflornithine is much smaller (+0.073) than that of ornithine (+0.99). Further results indicated that eflornithine utilized a saturable transport mechanism(s) to cross the hCMEC/D3 cell membranes and that transport was inhibited by the presence of other amino acids including ornithine. Eflornithine transport was also sodium-independent and sensitive to a y+-system inhibitor, but not a B0,+-system inhibitor. Eflornithine transport was also inhibited by pentamidine, suggestive of transport by organic cation transporters (OCT) which are expressed in this cell line. We confirmed expression of the y+-system protein, CAT1, and the B0,+-system protein, ATB0,+, in the hCMEC/D3 cells. We conclude that eflornithine uses the cationic amino acid transporter, system y+, and OCT to cross the BBB. This research highlights the potential of system-y+ to deliver drugs, including eflornithine, across the BBB to treat brain diseases.
Keywords: OCT; amino acids; blood-brain barrier; eflornithine; transporter; trypanosomiasis; y+-system.
Conflict of interest statement
Conflict of Interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Chemotherapy for second-stage human African trypanosomiasis.Cochrane Database Syst Rev. 2013 Jun 28;2013(6):CD006201. doi: 10.1002/14651858.CD006201.pub3. Cochrane Database Syst Rev. 2013. PMID: 23807762 Free PMC article.
-
Chemotherapy for second-stage Human African trypanosomiasis.Cochrane Database Syst Rev. 2010 Aug 4;(8):CD006201. doi: 10.1002/14651858.CD006201.pub2. Cochrane Database Syst Rev. 2010. Update in: Cochrane Database Syst Rev. 2013 Jun 28;(6):CD006201. doi: 10.1002/14651858.CD006201.pub3. PMID: 20687080 Updated.
-
Chemotherapy for second-stage human African trypanosomiasis: drugs in use.Cochrane Database Syst Rev. 2021 Dec 9;12(12):CD015374. doi: 10.1002/14651858.CD015374. Cochrane Database Syst Rev. 2021. PMID: 34882307 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
High-throughput drug screening to investigate blood-brain barrier permeability in vitro with a focus on breast cancer chemotherapeutic agents.Front Drug Deliv. 2024 Jun 27;4:1331126. doi: 10.3389/fddev.2024.1331126. eCollection 2024. Front Drug Deliv. 2024. PMID: 40836976 Free PMC article.
-
Saturation kinetics and specificity of transporters for L-arginine and asymmetric dimethylarginine (ADMA) at the blood-brain and blood-CSF barriers.PLoS One. 2025 Jun 11;20(6):e0320034. doi: 10.1371/journal.pone.0320034. eCollection 2025. PLoS One. 2025. PMID: 40498714 Free PMC article.
-
The microcirculation, the blood-brain barrier, and the neurovascular unit in health and Alzheimer disease: The aberrant pericyte is a central player.Pharmacol Rev. 2025 May;77(3):100052. doi: 10.1016/j.pharmr.2025.100052. Epub 2025 Mar 13. Pharmacol Rev. 2025. PMID: 40215558 Free PMC article. Review.
-
L-Arginine and asymmetric dimethylarginine (ADMA) transport across the mouse blood-brain and blood-CSF barriers: Evidence of saturable transport at both interfaces and CNS to blood efflux.PLoS One. 2024 Oct 24;19(10):e0305318. doi: 10.1371/journal.pone.0305318. eCollection 2024. PLoS One. 2024. PMID: 39446890 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

