OGDH and Bcl-xL loss causes synthetic lethality in glioblastoma
- PMID: 38483541
- PMCID: PMC11141877
- DOI: 10.1172/jci.insight.172565
OGDH and Bcl-xL loss causes synthetic lethality in glioblastoma
Abstract
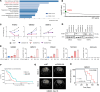
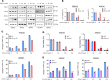
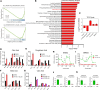
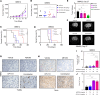
Glioblastoma (GBM) remains an incurable disease, requiring more effective therapies. Through interrogation of publicly available CRISPR and RNAi library screens, we identified the α-ketoglutarate dehydrogenase (OGDH) gene, which encodes an enzyme that is part of the tricarboxylic acid (TCA) cycle, as essential for GBM growth. Moreover, by combining transcriptome and metabolite screening analyses, we discovered that loss of function of OGDH by the clinically validated drug compound CPI-613 was synthetically lethal with Bcl-xL inhibition (genetically and through the clinically validated BH3 mimetic, ABT263) in patient-derived xenografts as well neurosphere GBM cultures. CPI-613-mediated energy deprivation drove an integrated stress response with an upregulation of the BH3-only domain protein, Noxa, in an ATF4-dependent manner, as demonstrated by genetic loss-of-function experiments. Consistently, silencing of Noxa attenuated cell death induced by CPI-613 in model systems of GBM. In patient-derived xenograft models of GBM in mice, the combination treatment of ABT263 and CPI-613 suppressed tumor growth and extended animal survival more potently than each compound on its own. Therefore, combined inhibition of Bcl-xL along with disruption of the TCA cycle might be a treatment strategy for GBM.
Keywords: Apoptosis pathways; Oncology.
Conflict of interest statement
Figures


References
-
- Bayik D, et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov. 2020;10(8):1210–1225. doi: 10.1158/2159-8290.CD-19-1355. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

