Frequency-potency analysis of IgG+ memory B cells delineates neutralizing antibody responses at single-cell resolution
- PMID: 38483908
- PMCID: PMC11003769
- DOI: 10.1016/j.celrep.2024.113948
Frequency-potency analysis of IgG+ memory B cells delineates neutralizing antibody responses at single-cell resolution
Abstract
Identifying individual functional B cell receptors (BCRs) is common, but two-dimensional analysis of B cell frequency versus BCR potency would delineate both quantity and quality of antigen-specific memory B cells. We efficiently determine quantitative BCR neutralizing activities using a single-cell-derived antibody supernatant analysis (SCAN) workflow and develop a frequency-potency algorithm to estimate B cell frequencies at various neutralizing activity or binding affinity cutoffs. In an HIV-1 fusion peptide (FP) immunization study, frequency-potency curves elucidate the quantity and quality of FP-specific immunoglobulin G (IgG)+ memory B cells for different animals, time points, and antibody lineages at single-cell resolution. The BCR neutralizing activities are mainly determined by their affinities to soluble envelope trimer. Frequency analysis definitively demonstrates dominant neutralizing antibody lineages. These findings establish SCAN and frequency-potency analyses as promising approaches for general B cell analysis and monoclonal antibody (mAb) discovery. They also provide specific rationales for HIV-1 FP-directed vaccine optimization.
Keywords: B cell receptor; BCR; CP: Immunology; HIV-1; SCAN; antibody; frequency; fusion peptide; potency; single-cell-derived antibody supernatant analysis; vaccine.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




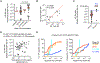
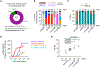
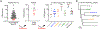
References
-
- Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, Gillespie RA, Ransier A, Darko S, Hu J, et al. (2019). Activation Dynamics and Immunoglobulin Evolution of Pre-existing and Newly Generated Human Memory B cell Responses to Influenza Hemagglutinin. Immunity 51, 398–410.e5. 10.1016/j.immuni.2019.06.024. - DOI - PubMed
-
- Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, et al. (2021). First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci. Transl. Med 13, eabg4535. 10.1126/scitranslmed.abg4535. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

