The polyglutamine domain is the primary driver of seeding in huntingtin aggregation
- PMID: 38483973
- PMCID: PMC10939245
- DOI: 10.1371/journal.pone.0298323
The polyglutamine domain is the primary driver of seeding in huntingtin aggregation
Abstract
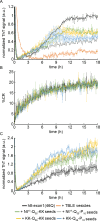
Huntington's Disease (HD) is a fatal, neurodegenerative disease caused by aggregation of the huntingtin protein (htt) with an expanded polyglutamine (polyQ) domain into amyloid fibrils. Htt aggregation is modified by flanking sequences surrounding the polyQ domain as well as the binding of htt to lipid membranes. Upon fibrillization, htt fibrils are able to template the aggregation of monomers into fibrils in a phenomenon known as seeding, and this process appears to play a critical role in cell-to-cell spread of HD. Here, exposure of C. elegans expressing a nonpathogenic N-terminal htt fragment (15-repeat glutamine residues) to preformed htt-exon1 fibrils induced inclusion formation and resulted in decreased viability in a dose dependent manner, demonstrating that seeding can induce toxic aggregation of nonpathogenic forms of htt. To better understand this seeding process, the impact of flanking sequences adjacent to the polyQ stretch, polyQ length, and the presence of model lipid membranes on htt seeding was investigated. Htt seeding readily occurred across polyQ lengths and was independent of flanking sequence, suggesting that the structured polyQ domain within fibrils is the key contributor to the seeding phenomenon. However, the addition of lipid vesicles modified seeding efficiency in a manner suggesting that seeding primarily occurs in bulk solution and not at the membrane interface. In addition, fibrils formed in the presence of lipid membranes displayed similar seeding efficiencies. Collectively, this suggests that the polyQ domain that forms the amyloid fibril core is the main driver of seeding in htt aggregation.
Copyright: © 2024 Skeens et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- The Collaborative Huntingtons Disease Research Group. A novel gene containing a trinucleotide repeat that is exanded and unstable on Huntingtons-disease chromosomes Cell. 1993;72(6):971–83. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

