Analytical validation of NeXT Personal®, an ultra-sensitive personalized circulating tumor DNA assay
- PMID: 38484152
- PMCID: PMC10939476
- DOI: 10.18632/oncotarget.28565
Analytical validation of NeXT Personal®, an ultra-sensitive personalized circulating tumor DNA assay
Abstract
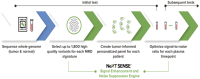
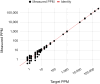
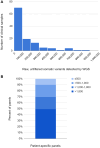
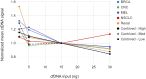
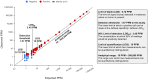
We describe the analytical validation of NeXT Personal®, an ultra-sensitive, tumor-informed circulating tumor DNA (ctDNA) assay for detecting residual disease, monitoring therapy response, and detecting recurrence in patients diagnosed with solid tumor cancers. NeXT Personal uses whole genome sequencing of tumor and matched normal samples combined with advanced analytics to accurately identify up to ~1,800 somatic variants specific to the patient's tumor. A personalized panel is created, targeting these variants and then used to sequence cell-free DNA extracted from patient plasma samples for ultra-sensitive detection of ctDNA. The NeXT Personal analytical validation is based on panels designed from tumor and matched normal samples from two cell lines, and from 123 patients across nine cancer types. Analytical measurements demonstrated a detection threshold of 1.67 parts per million (PPM) with a limit of detection at 95% (LOD95) of 3.45 PPM. NeXT Personal showed linearity over a range of 0.8 to 300,000 PPM (Pearson correlation coefficient = 0.9998). Precision varied from a coefficient of variation of 12.8% to 3.6% over a range of 25 to 25,000 PPM. The assay targets 99.9% specificity, with this validation study measuring 100% specificity and in silico methods giving us a confidence interval of 99.92 to 100%. In summary, this study demonstrates NeXT Personal as an ultra-sensitive, highly quantitative and robust ctDNA assay that can be used to detect residual disease, monitor treatment response, and detect recurrence in patients.
Keywords: analytical validation; circulating tumor DNA; molecular residual disease; tumor-informed assay; whole genome sequencing.
Conflict of interest statement
All authors have or had a financial relationship as employees of Personalis, Inc.
Figures



References
-
- Ococks E, Frankell AM, Masque Soler N, Grehan N, Northrop A, Coles H, Redmond AM, Devonshire G, Weaver JMJ, Hughes C, Lehovsky K, Blasko A, Nutzinger B, et al.. Longitudinal tracking of 97 esophageal adenocarcinomas using liquid biopsy sampling. Ann Oncol. 2021; 32:522–32. 10.1016/j.annonc.2020.12.010. - DOI - PubMed
-
- Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL, Zhou L, Scherer F, Kurtz DM, Say C, et al.. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017; 7:1394–403. 10.1158/2159-8290.CD-17-0716. - DOI - PMC - PubMed
-
- Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, Ali S, Cleator S, Kenny L, Stebbing J, Rutherford M, Sethi H, Boydell A, et al.. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res. 2019; 25:4255–63. 10.1158/1078-0432.CCR-18-3663. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

