Genomes in clinical care
- PMID: 38485733
- PMCID: PMC10940576
- DOI: 10.1038/s41525-024-00402-2
Genomes in clinical care
Abstract
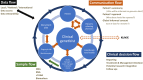
In the era of precision medicine, genome sequencing (GS) has become more affordable and the importance of genomics and multi-omics in clinical care is increasingly being recognized. However, how to scale and effectively implement GS on an institutional level remains a challenge for many. Here, we present Genome First and Ge-Med, two clinical implementation studies focused on identifying the key pillars and processes that are required to make routine GS and predictive genomics a reality in the clinical setting. We describe our experience and lessons learned for a variety of topics including test logistics, patient care processes, data reporting, and infrastructure. Our model of providing clinical care and comprehensive genomic analysis from a single source may be used by other centers with a similar structure to facilitate the implementation of omics-based personalized health concepts in medicine.
© 2024. The Author(s).
Conflict of interest statement
S.T. and S.S. declare to be employees and shareholders of Illumina, Inc. S.O. received travel support and speaker fees from Illumina, Inc. and Oxford Nanopore Technologies, Inc. O.R., T.H., S.O., and C.S. report institutional grants from Illumina, and C.S. has received research grants from BMS Stiftung Immunonkologie and Westdeutsche Studiengruppe GmbH outside the submitted work. All other authors declare no competing interests.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources

