Enterobacter asburiae ST229: an emerging carbapenemases producer
- PMID: 38486043
- PMCID: PMC10940580
- DOI: 10.1038/s41598-024-55884-y
Enterobacter asburiae ST229: an emerging carbapenemases producer
Erratum in
-
Publisher Correction: Enterobacter asburiae ST229: an emerging carbapenemases producer.Sci Rep. 2024 May 1;14(1):9987. doi: 10.1038/s41598-024-60104-8. Sci Rep. 2024. PMID: 38693199 Free PMC article. No abstract available.
Abstract
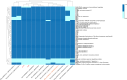
Enterobacter asburiae, member of the Enterobacter cloacae complex (ECC) group, shows an increasing clinical relevance being responsible for infections like pneumonia, urinary tract infections and septicemia. The aim of the present study was the investigation of the genomic features of two XDR E. asburiae ST229 clinical strains co-carrying blaNDM-1 and blaVIM-1 determinants, collected in October 2021 and in June 2022, respectively. Two E. asburiae strains were collected from rectal swabs of as many patients admitted to the cardiopulmonary intensive care unit of Fondazione I.R.C.C.S. "Policlinico San Matteo" in Pavia, Italy. Based on the antibiotic susceptibility profile results, both isolates showed an XDR phenotype, retaining susceptibility only to fluoroquinolones. Both isolates shared identical resistome, virulome, plasmid content, and belonged to ST229, a rarely reported sequence type. They co-harbored blaNDM-1 and blaVIM-1 genes, that resulted located on transferable plasmids by conjugation and transformation. Moreover, both strains differed in 24 SNPs and showed genetic relatedness with E. asburiae ST709 and ST27. We described the first case of ST229 E. asburiae co-harboring blaNDM-1 and blaVIM-1 in Italy. This study points out the emergence of carbapenemases in low-risk pathogens, representing a novel challenge for public health, that should include such types of strains in dedicated surveillance programs. Antimicrobial susceptibility testing was carried out using Thermo Scientific™ Sensititre™ Gram Negative MIC Plates DKMGN. Both strains underwent whole-genome sequencing (WGS) using Illumina Miseq platform. Resistome, plasmidome, virulome, MLST, plasmid MLST and a SNPs-based phylogenetic tree were in silico determined.
Keywords: Enterobacter asburiae; Carbapenemase-producing; Epidemiology; Low-risk pathogens; NDM-1; Surveillance; VIM-1; WGS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
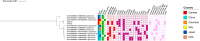
References
-
- Huang Z, Liu S, Wang Y, Yao Z, Feng L, Lin Y, Ye J, Zhou T, Wang Z. Comparison of prevalence, resistance, biofilm-forming ability and virulence between carbapenem-non-susceptible and carbepenem-susceptible Enterobacter cloacae complex in clusters. J. Hosp. Infect. 2023;139:168–174. doi: 10.1016/j.jhin.2023.06.017. - DOI - PubMed
-
- Candela A, Guerrero-López A, Mateos M, Gómez-Asenjo A, Arroyo MJ, Hernandez-García M, Del Campo R, Cercenado E, Cuénod A, Méndez G, Mancera L, Caballero JD, Martínez-García L, Gijón D, Morosini MI, Ruiz-Garbajosa P, Egli A, Cantón R, Muñoz P, Rodríguez-Temporal D, Rodríguez-Sánchez B. Automatic discrimination of species within the Enterobacter cloacae complex using matrix-assisted laser desorption ionization-time of flight mass spectrometry and supervised algorithms. J. Clin. Microbiol. 2023;61(4):e0104922. doi: 10.1128/jcm.01049-22. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

