A neural network-based model framework for cell-fate decisions and development
- PMID: 38486083
- PMCID: PMC10940658
- DOI: 10.1038/s42003-024-05985-1
A neural network-based model framework for cell-fate decisions and development
Abstract
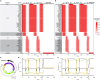
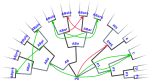
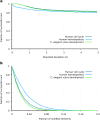
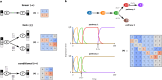
Gene regulatory networks (GRNs) fulfill the essential function of maintaining the stability of cellular differentiation states by sustaining lineage-specific gene expression, while driving the progression of development. However, accounting for the relative stability of intermediate differentiation stages and their divergent trajectories remains a major challenge for models of developmental biology. Here, we develop an empirical data-based associative GRN model (AGRN) in which regulatory networks store multilineage stage-specific gene expression profiles as associative memory patterns. These networks are capable of responding to multiple instructive signals and, depending on signal timing and identity, can dynamically drive the differentiation of multipotent cells toward different cell state attractors. The AGRN dynamics can thus generate diverse lineage-committed cell populations in a robust yet flexible manner, providing an attractor-based explanation for signal-driven cell fate decisions during differentiation and offering a readily generalizable modelling tool that can be applied to a wide variety of cell specification systems.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

