Cancer-derived exosomal Alu RNA promotes colorectal cancer progression
- PMID: 38486106
- PMCID: PMC10984964
- DOI: 10.1038/s12276-024-01166-6
Cancer-derived exosomal Alu RNA promotes colorectal cancer progression
Abstract
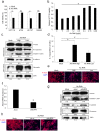
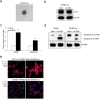
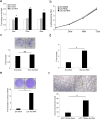
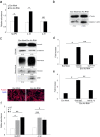
Inflammation plays a crucial role in cancer progression, but the relevance of the inflammasome remains unclear. Alu RNA was the first endogenous nucleic acid shown to activate the NLRP3 (nucleotide-binding domain leucine-rich repeat containing 3) inflammasome. Here, we showed that Alu RNA can induce epithelial-to-mesenchymal transition (EMT) through NLRP3 inflammasome activation and IL-1β release in colorectal cancer (CRC) cells. Alu RNA is stored, transported and transferred to CRC cells by exosomes. Exosomal Alu RNA promotes tumorigenesis by inducing invasion, metastasis and EMT via NLRP3 inflammasome activation. Consistent with these data, we found that significantly increased Alu RNA expression correlates with the induction of NLRP3 priming in human CRC patients. Furthermore, the level of Alu RNA in circulating exosomes correlates with CRC progression in a preclinical model. These findings reveal the direct involvement of Alu RNA in cancer pathogenesis, and its presence in CRC cell-derived exosomes could be used as a noninvasive diagnostic biomarker.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

