NFKBIE mutations are selected by the tumor microenvironment and contribute to immune escape in chronic lymphocytic leukemia
- PMID: 38486128
- PMCID: PMC11216988
- DOI: 10.1038/s41375-024-02224-8
NFKBIE mutations are selected by the tumor microenvironment and contribute to immune escape in chronic lymphocytic leukemia
Abstract
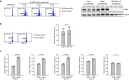
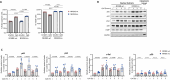
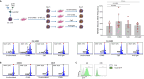
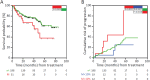
Loss-of-function mutations in NFKBIE, which encodes for the NF-κB inhibitor IκBε, are frequent in chronic lymphocytic leukemia (CLL) and certain other B-cell malignancies and have been associated with accelerated disease progression and inferior responses to chemotherapy. Using in vitro and in vivo murine models and primary patient samples, we now show that NFKBIE-mutated CLL cells are selected by microenvironmental signals that activate the NF-κB pathway and induce alterations within the tumor microenvironment that can allow for immune escape, including expansion of CD8+ T-cells with an exhausted phenotype and increased PD-L1 expression on the malignant B-cells. Consistent with the latter observations, we find increased expression of exhaustion markers on T-cells from patients with NFKBIE-mutated CLL. In addition, we show that NFKBIE-mutated murine CLL cells display selective resistance to ibrutinib and report inferior outcomes to ibrutinib treatment in NFKBIE-mutated CLL patients. These findings suggest that NFKBIE mutations can contribute to CLL progression through multiple mechanisms, including a bidirectional crosstalk with the microenvironment and reduced sensitivity to BTK inhibitor treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

