XX sex chromosome complement modulates immune responses to heat-killed Streptococcus pneumoniae immunization in a microbiome-dependent manner
- PMID: 38486287
- PMCID: PMC10938708
- DOI: 10.1186/s13293-024-00597-0
XX sex chromosome complement modulates immune responses to heat-killed Streptococcus pneumoniae immunization in a microbiome-dependent manner
Abstract
Background: Differences in male vs. female immune responses are well-documented and have significant clinical implications. While the immunomodulatory effects of sex hormones are well established, the contributions of sex chromosome complement (XX vs. XY) and gut microbiome diversity on immune sexual dimorphisms have only recently become appreciated. Here we investigate the individual and collaborative influences of sex chromosome complements and gut microbiota on humoral immune activation.
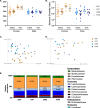
Methods: Male and female Four Core Genotype (FCG) mice were immunized with heat-killed Streptococcus pneumoniae (HKSP). Humoral immune responses were assessed, and X-linked immune-related gene expression was evaluated to explain the identified XX-dependent phenotype. The functional role of Kdm6a, an X-linked epigenetic regulatory gene of interest, was evaluated ex vivo using mitogen stimulation of B cells. Additional influences of the gut microbiome on sex chromosome-dependent B cell activation was also evaluated by antibiotically depleting gut microbiota prior to HKSP immunization. Reconstitution of the depleted microbiome with short-chain fatty acid (SCFA)-producing bacteria tested the impact of SCFAs on XX-dependent immune activation.
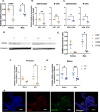
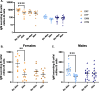
Results: XX mice exhibited higher HKSP-specific IgM-secreting B cells and plasma cell frequencies than XY mice, regardless of gonadal sex. Although Kdm6a was identified as an X-linked gene overexpressed in XX B cells, inhibition of its enzymatic activity did not affect mitogen-induced plasma cell differentiation or antibody production in a sex chromosome-dependent manner ex vivo. Enhanced humoral responses in XX vs. XY immunized FCG mice were eliminated after microbiome depletion, indicating that the microbiome contributes to the identified XX-dependent immune enhancement. Reconstituting microbiota-depleted mice with select SCFA-producing bacteria enhanced fecal SCFA concentrations and increased humoral responses in XX, but not XY, FCG mice. However, exposure to the SCFA propionate alone did not enhance mitogenic B cell stimulation in ex vivo studies.
Conclusions: FCG mice have been used to assess sex hormone and sex chromosome complement influences on various sexually dimorphic traits. The current study indicates that the gut microbiome impacts humoral responses in an XX-dependent manner, suggesting that the collaborative influence of gut bacteria and other sex-specific factors should be considered when interpreting data aimed at delineating the mechanisms that promote sexual dimorphism.
Keywords: Four Core Genotype; Gut microbiome; HKSP; IgM; Kdm6a; Plasma cells; SCFA; Sex differences; X chromosome.
Plain language summary
Male and female immune systems differ in their ability to respond to infectious challenge. While males tend to be more susceptible to infection and produce lower amounts of antibodies in response to vaccination, females are more prone to develop autoimmune and inflammatory diseases. Key contributors to these differences include sex hormones, sex chromosome complement (XX in females vs. XY in males), and distinct gut microbial communities capable of regulating immune activation. While each factor has been studied individually, this research underscores the potential for these factors to collaboratively impact immune activation. Here, possession of an XX vs. XY sex chromosome complement was demonstrated to enhance antibody responses to heat-killed Streptococcus pneumoniae vaccination. While attempting to determine the underlying cause of this immune enhancement, the gut microbiome was identified to play a critical role. In the absence of an intact gut microbiome, XX immune activation was reduced to levels similar to those seen in XY sex chromosome complement-possessing mice. Replacement of the depleted gut microbiomes with select SCFA-producing bacterial species enhanced SCFA levels in antibiotic-treated mice and rescued the XX-dependent immune enhancement, suggesting a SCFA-mediated contribution. Further studies are needed to determine exactly how these select bacteria impact immune activation in a sex chromosome complement-dependent manner. Our findings highlight the need to consider the collaborative effects of individual sex-specific factors when attempting to understand immune sex biases, as a better understanding of these interactions will likely pave the way for improving therapeutics and vaccines tailored to both sexes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Update of
-
XX sex chromosome complement modulates immune responses to heat-killed Streptococcus pneumoniae immunization in a microbiome-dependent manner.Res Sq [Preprint]. 2023 Nov 2:rs.3.rs-3429829. doi: 10.21203/rs.3.rs-3429829/v1. Res Sq. 2023. Update in: Biol Sex Differ. 2024 Mar 14;15(1):21. doi: 10.1186/s13293-024-00597-0. PMID: 37961596 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

