Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection
- PMID: 38486802
- PMCID: PMC10939363
- DOI: 10.1093/ve/veae001
Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection
Abstract
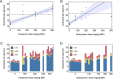
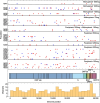
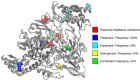
Mutagenic antiviral drugs have shown promise against multiple viruses, but concerns have been raised about whether their use might promote the emergence of new and harmful viral variants. Recently, genetic signatures associated with molnupiravir use have been identified in the global SARS-COV-2 population. Here, we examine the consequences of using favipiravir and molnupiravir to treat SARS-CoV-2 infection in a hamster model, comparing viral genome sequence data collected from (1) untreated hamsters, and (2) from hamsters receiving effective and suboptimal doses of treatment. We identify a broadly linear relationship between drug dose and the extent of variation in treated viral populations, with a high proportion of this variation being composed of variants at frequencies of less than 1 per cent, below typical thresholds for variant calling. Treatment with an effective dose of antiviral drug was associated with a gain of between 7 and 10 variants per viral genome relative to drug-free controls: even after a short period of treatment a population founded by a transmitted virus could contain multiple sequence differences to that of the original host. Treatment with a suboptimal dose of drug showed intermediate gains of variants. No dose-dependent signal was identified in the numbers of single-nucleotide variants reaching frequencies in excess of 5 per cent. We did not find evidence to support the emergence of drug resistance or of novel immune phenotypes. Our study suggests that where onward transmission occurs, a short period of treatment with mutagenic drugs may be sufficient to generate a significant increase in the number of viral variants transmitted.
Keywords: SARS-CoV-2; favipiravir; molnupiravir; mutagenesis.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

