Detection of Patients at Risk of Multidrug-Resistant Enterobacteriaceae Infection Using Graph Neural Networks: A Retrospective Study
- PMID: 38487204
- PMCID: PMC10904075
- DOI: 10.34133/hds.0099
Detection of Patients at Risk of Multidrug-Resistant Enterobacteriaceae Infection Using Graph Neural Networks: A Retrospective Study
Erratum in
-
Erratum to "Detection of Patients at Risk of Multidrug-Resistant Enterobacteriaceae Infection Using Graph Neural Networks: A Retrospective Study".Health Data Sci. 2023 Dec 16;4:0216. doi: 10.34133/hds.0216. eCollection 2024. Health Data Sci. 2023. PMID: 39687411 Free PMC article.
Abstract
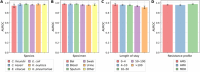
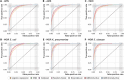
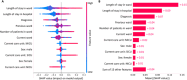
Background: While Enterobacteriaceae bacteria are commonly found in the healthy human gut, their colonization of other body parts can potentially evolve into serious infections and health threats. We investigate a graph-based machine learning model to predict risks of inpatient colonization by multidrug-resistant (MDR) Enterobacteriaceae. Methods: Colonization prediction was defined as a binary task, where the goal is to predict whether a patient is colonized by MDR Enterobacteriaceae in an undesirable body part during their hospital stay. To capture topological features, interactions among patients and healthcare workers were modeled using a graph structure, where patients are described by nodes and their interactions are described by edges. Then, a graph neural network (GNN) model was trained to learn colonization patterns from the patient network enriched with clinical and spatiotemporal features. Results: The GNN model achieves performance between 0.91 and 0.96 area under the receiver operating characteristic curve (AUROC) when trained in inductive and transductive settings, respectively, up to 8% above a logistic regression baseline (0.88). Comparing network topologies, the configuration considering ward-related edges (0.91 inductive, 0.96 transductive) outperforms the configurations considering caregiver-related edges (0.88, 0.89) and both types of edges (0.90, 0.94). For the top 3 most prevalent MDR Enterobacteriaceae, the AUROC varies from 0.94 for Citrobacter freundii up to 0.98 for Enterobacter cloacae using the best-performing GNN model. Conclusion: Topological features via graph modeling improve the performance of machine learning models for Enterobacteriaceae colonization prediction. GNNs could be used to support infection prevention and control programs to detect patients at risk of colonization by MDR Enterobacteriaceae and other bacteria families.
Copyright © 2023 Racha Gouareb et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures




References
-
- Allegranzi B, Nejad SB, Combescure C, Graafmans W, Attar H, Donaldson L, Pittet D. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. - PubMed
-
- World Health Organization. Charter: Health worker safety: A priority for patient safety. Geneva (Switzerland): World Health Organization; 2020.
-
- World Health Organization. Report on the burden of endemic health care-associated infection worldwide. Geneva (Switzerland): World Health Organization; 2011.
-
- Patient Carelink. Healthcare-acquired infections (HAIs). 2022. Available at http://patientcarelink.org/improving-patient-care/healthcare-acquired-in... [accessed October 10, 2022].
LinkOut - more resources
Full Text Sources

