FLUTE: A Python GUI for interactive phasor analysis of FLIM data
- PMID: 38487690
- PMCID: PMC10936343
- DOI: 10.1017/S2633903X23000211
FLUTE: A Python GUI for interactive phasor analysis of FLIM data
Abstract
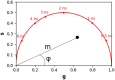
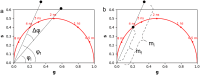
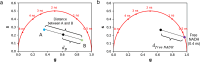
Fluorescence lifetime imaging microscopy (FLIM) is a powerful technique used to probe the local environment of fluorophores. The fit-free phasor approach to FLIM data is increasingly being used due to its ease of interpretation. To date, no open-source graphical user interface (GUI) for phasor analysis of FLIM data is available in Python, thus limiting the widespread use of phasor analysis in biomedical research. Here, we present Fluorescence Lifetime Ultimate Explorer (FLUTE), a Python GUI that is designed to fill this gap. FLUTE simplifies and automates many aspects of the analysis of FLIM data acquired in the time domain, such as calibrating the FLIM data, performing interactive exploration of the phasor plot, displaying phasor plots and FLIM images with different lifetime contrasts simultaneously, and calculating the distance from known molecular species. After applying desired filters and thresholds, the final edited datasets can be exported for further user-specific analysis. FLUTE has been tested using several FLIM datasets including autofluorescence of zebrafish embryos and in vitro cells. In summary, our user-friendly GUI extends the advantages of phasor plotting by making the data visualization and analysis easy and interactive, allows for analysis of large FLIM datasets, and accelerates FLIM analysis for non-specialized labs.
Keywords: Data visualization; FLIM; Python; fluorescence lifetime microscopy; phasor analysis.
© The Author(s) 2023.
Conflict of interest statement
The authors declare no competing interests exist.
Figures






References
-
- Wallrabe H & Periasamy A (2005) Imaging protein molecules using FRET and FLIM microscopy. Current Opinion in Biotechnology 16(1), 19–27. - PubMed
-
- Becker W (2012) Fluorescence lifetime imaging – techniques and applications. Journal of Microscopy 247(2), 119–136. - PubMed
-
- Suhling K, Hirvonen LM, Levitt JA, Chung PH, Tregidgo C, le Marois A, Rusakov DA, Zheng K, Ameer-Beg S, Poland S, Coelho S, Henderson R & Krstajic N (2015) Fluorescence lifetime imaging (FLIM): Basic concepts and some recent developments. Medical Photonics 27, 3–40.
LinkOut - more resources
Full Text Sources
Miscellaneous
