Federated learning for multi-omics: A performance evaluation in Parkinson's disease
- PMID: 38487808
- PMCID: PMC10935499
- DOI: 10.1016/j.patter.2024.100945
Federated learning for multi-omics: A performance evaluation in Parkinson's disease
Abstract
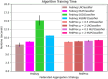
While machine learning (ML) research has recently grown more in popularity, its application in the omics domain is constrained by access to sufficiently large, high-quality datasets needed to train ML models. Federated learning (FL) represents an opportunity to enable collaborative curation of such datasets among participating institutions. We compare the simulated performance of several models trained using FL against classically trained ML models on the task of multi-omics Parkinson's disease prediction. We find that FL model performance tracks centrally trained ML models, where the most performant FL model achieves an AUC-PR of 0.876 ± 0.009, 0.014 ± 0.003 less than its centrally trained variation. We also determine that the dispersion of samples within a federation plays a meaningful role in model performance. Our study implements several open-source FL frameworks and aims to highlight some of the challenges and opportunities when applying these collaborative methods in multi-omics studies.
Keywords: Parkinson’s disease diagnosis; federated learning; machine learning; omics data analysis.
© 2024 The Authors.
Conflict of interest statement
B.P.D., A.D., D.V., M.A.N., and F.F. declare the following competing financial interests, as their participation in this project was part of a competitive contract awarded to Data Tecnica LLC by the National Institutes of Health to support open science research. M.A.N. also currently serves on the scientific advisory board for Character Bio and is an advisor to Neuron23 Inc. The study’s funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. F.F. takes final responsibility for the decision to submit the paper for publication.
Figures





Update of
-
Federated Learning for multi-omics: a performance evaluation in Parkinson's disease.bioRxiv [Preprint]. 2024 Feb 12:2023.10.04.560604. doi: 10.1101/2023.10.04.560604. bioRxiv. 2024. Update in: Patterns (N Y). 2024 Mar 01;5(3):100945. doi: 10.1016/j.patter.2024.100945. PMID: 37986893 Free PMC article. Updated. Preprint.
References
-
- Dadu A., Satone V., Kaur R., Hashemi S.H., Leonard H., Iwaki H., Makarious M.B., Billingsley K.J., Bandres-Ciga S., Sargent L.J., et al. Identification and prediction of Parkinson’s disease subtypes and progression using machine learning in two cohorts. NPJ Parkinsons Dis. 2022;8:172. doi: 10.1038/s41531-022-00439-z. - DOI - PMC - PubMed
-
- Green E.D., Gunter C., Biesecker L.G., Di Francesco V., Easter C.L., Feingold E.A., Felsenfeld A.L., Kaufman D.J., Ostrander E.A., Pavan W.J., et al. Strategic vision for improving human health at The Forefront of Genomics. Nature. 2020;586:683–692. doi: 10.1038/s41586-020-2817-4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

