Manipulation of Glutamatergic Neuronal Activity in the Primary Motor Cortex Regulates Cardiac Function in Normal and Myocardial Infarction Mice
- PMID: 38488323
- PMCID: PMC11132081
- DOI: 10.1002/advs.202305581
Manipulation of Glutamatergic Neuronal Activity in the Primary Motor Cortex Regulates Cardiac Function in Normal and Myocardial Infarction Mice
Abstract
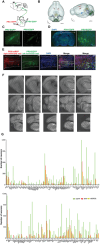
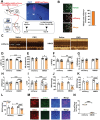
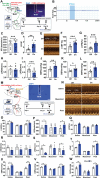
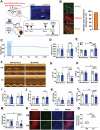
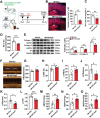
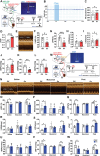
Cardiac function is under neural regulation; however, brain regions in the cerebral cortex responsible for regulating cardiac function remain elusive. In this study, retrograde trans-synaptic viral tracing is used from the heart to identify a specific population of the excitatory neurons in the primary motor cortex (M1) that influences cardiac function in mice. Optogenetic activation of M1 glutamatergic neurons increases heart rate, ejection fraction, and blood pressure. By contrast, inhibition of M1 glutamatergic neurons decreased cardiac function and blood pressure as well as tyrosine hydroxylase (TH) expression in the heart. Using viral tracing and optogenetics, the median raphe nucleus (MnR) is identified as one of the key relay brain regions in the circuit from M1 that affect cardiac function. Then, a mouse model of cardiac injury is established caused by myocardial infarction (MI), in which optogenetic activation of M1 glutamatergic neurons impaired cardiac function in MI mice. Moreover, ablation of M1 neurons decreased the levels of norepinephrine and cardiac TH expression, and enhanced cardiac function in MI mice. These findings establish that the M1 neurons involved in the regulation of cardiac function and blood pressure. They also help the understanding of the neural mechanisms underlying cardiovascular regulation.
Keywords: cardiac function; median raphe nuclei; myocardial infarction; primary motor cortex.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- a) Tahsili‐Fahadan P., Geocadin R. G., Circ. Res. 2017, 120, 559; - PubMed
- b) Shivkumar K., Ajijola O. A., Anand I., Armour J. A., Chen P. S., Esler M., De Ferrari G. M., Fishbein M. C., Goldberger J. J., Harper R. M., Joyner M. J., Khalsa S. S., Kumar R., Lane R., Mahajan A., Po S., Schwartz P. J., Somers V. K., Valderrabano M., Vaseghi M., Zipes D. P., J. Physiol. 2016, 594, 3911; - PMC - PubMed
- c) Kataoka N., Shima Y., Nakajima K., Nakamura K., Science 2020, 367, 1105; - PubMed
- d) Sanchez‐Larsen A., Principe A., Ley M., Navarro‐Cuartero J., Rocamora R., Ann. Neurol. 2021, 89, 1172; - PubMed
-
- Ter Horst G. J., Postema F., Am. J. Physiol. 1997, 273, H2926,. - PubMed
-
- Hsueh B., Chen R., Jo Y., Tang D., Raffiee M., Kim Y. S., Inoue M., Randles S., Ramakrishnan C., Patel S., Kim D. K., Liu T. X., Kim S. H., Tan L., Mortazavi L., Cordero A., Shi J., Zhao M., Ho T. T., Crow A., Yoo A. W., Raja C., Evans K., Bernstein D., Zeineh M., Goubran M., Deisseroth K., Nature 2023, 615, 292. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
