Mitochondrial genomes of Pleistocene megafauna retrieved from recent sediment layers of two Siberian lakes
- PMID: 38488477
- PMCID: PMC10942779
- DOI: 10.7554/eLife.89992
Mitochondrial genomes of Pleistocene megafauna retrieved from recent sediment layers of two Siberian lakes
Abstract
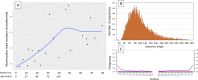
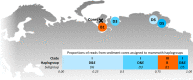
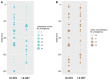
Ancient environmental DNA (aeDNA) from lake sediments has yielded remarkable insights for the reconstruction of past ecosystems, including suggestions of late survival of extinct species. However, translocation and lateral inflow of DNA in sediments can potentially distort the stratigraphic signal of the DNA. Using three different approaches on two short lake sediment cores of the Yamal peninsula, West Siberia, with ages spanning only the past hundreds of years, we detect DNA and identified mitochondrial genomes of multiple mammoth and woolly rhinoceros individuals-both species that have been extinct for thousands of years on the mainland. The occurrence of clearly identifiable aeDNA of extinct Pleistocene megafauna (e.g. >400 K reads in one core) throughout these two short subsurface cores, along with specificities of sedimentology and dating, confirm that processes acting on regional scales, such as extensive permafrost thawing, can influence the aeDNA record and should be accounted for in aeDNA paleoecology.
Keywords: genetics; genomics; mammal; mammoth; woolly rhinoceros.
© 2023, Seeber et al.
Conflict of interest statement
PS Employed by and owns stock of Thermo Fisher Scientific, LB, YD, AS, YW, KS, KM, BS, LE No competing interests declared, SV Employed by Embark Veterinary, Inc
Figures
Update of
- doi: 10.1101/2023.06.16.545324
- doi: 10.7554/eLife.89992.1
- doi: 10.7554/eLife.89992.2
References
-
- Alsos IG, Sjögren P, Brown AG, Gielly L, Merkel MKF, Paus A, Lammers Y, Edwards ME, Alm T, Leng M, Goslar T, Langdon CT, Bakke J, van der Bilt WGM. Last Glacial Maximum environmental conditions at Andøya, northern Norway; evidence for a northern ice-edge ecological “hotspot.”. Quaternary Science Reviews. 2020;239:106364. doi: 10.1016/j.quascirev.2020.106364. - DOI
-
- Armbrecht LH, Coolen MJL, Lejzerowicz F, George SC, Negandhi K, Suzuki Y, Young J, Foster NR, Armand LK, Cooper A, Ostrowski M, Focardi A, Stat M, Moreau JW, Weyrich LS. Ancient DNA from marine sediments: Precautions and considerations for seafloor coring, sample handling and data generation. Earth-Science Reviews. 2019;196:102887. doi: 10.1016/j.earscirev.2019.102887. - DOI
-
- Arnold LJ, Roberts RG, Macphee RDE, Haile JS, Brock F, Möller P, Froese DG, Tikhonov AN, Chivas AR, Gilbert MTP, Willerslev E. Paper II - Dirt, dates and DNA: OSL and radiocarbon chronologies of perennially frozen sediments in Siberia, and their implications for sedimentary ancient DNA studies. Boreas. 2011;40:417–445. doi: 10.1111/j.1502-3885.2010.00181.x. - DOI

