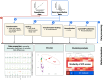
Data processing solutions to render metabolomics more quantitative: case studies in food and clinical metabolomics using Metabox 2.0
- PMID: 38488666
- PMCID: PMC10941642
- DOI: 10.1093/gigascience/giae005
Data processing solutions to render metabolomics more quantitative: case studies in food and clinical metabolomics using Metabox 2.0
Abstract
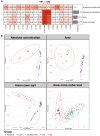
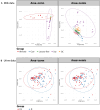
In classic semiquantitative metabolomics, metabolite intensities are affected by biological factors and other unwanted variations. A systematic evaluation of the data processing methods is crucial to identify adequate processing procedures for a given experimental setup. Current comparative studies are mostly focused on peak area data but not on absolute concentrations. In this study, we evaluated data processing methods to produce outputs that were most similar to the corresponding absolute quantified data. We examined the data distribution characteristics, fold difference patterns between 2 metabolites, and sample variance. We used 2 metabolomic datasets from a retail milk study and a lupus nephritis cohort as test cases. When studying the impact of data normalization, transformation, scaling, and combinations of these methods, we found that the cross-contribution compensating multiple standard normalization (ccmn) method, followed by square root data transformation, was most appropriate for a well-controlled study such as the milk study dataset. Regarding the lupus nephritis cohort study, only ccmn normalization could slightly improve the data quality of the noisy cohort. Since the assessment accounted for the resemblance between processed data and the corresponding absolute quantified data, our results denote a helpful guideline for processing metabolomic datasets within a similar context (food and clinical metabolomics). Finally, we introduce Metabox 2.0, which enables thorough analysis of metabolomic data, including data processing, biomarker analysis, integrative analysis, and data interpretation. It was successfully used to process and analyze the data in this study. An online web version is available at http://metsysbio.com/metabox.
Keywords: R package; data processing; metabolomics; normalization; quantitative analysis; scaling; semiquantitative analysis; transformation.
© The Author(s) 2024. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures