Global analysis of T-cell groups reveals immunological features and common antigen targets of digestive tract tumors
- PMID: 38488909
- PMCID: PMC10943170
- DOI: 10.1007/s00432-024-05645-1
Global analysis of T-cell groups reveals immunological features and common antigen targets of digestive tract tumors
Abstract
Background: T cells are key players in the tumor immune microenvironment (TIME), as they can recognize and eliminate cancer cells that express neoantigens derived from somatic mutations. However, the diversity and specificity of T-cell receptors (TCRs) that recognize neoantigens are largely unknown, due to the high variability of TCR sequences among individuals.
Methods: To address this challenge, we applied GLIPH2, a novel algorithm that groups TCRs based on their predicted antigen specificity and HLA restriction, to cluster the TCR repertoire of 1,702 patients with digestive tract cancer. The patients were divided into five groups based on whether they carried tumor-infiltrating or clonal-expanded TCRs and calculated their TCR diversity. The prognosis, tumor subtype, gene mutation, gene expression, and immune microenvironment of these groups were compared. Viral specificity inference and immunotherapy relevance analysis performed for the TCR groups.
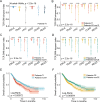
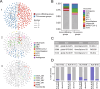
Results: This approach reduced the complexity of TCR sequences to 249 clonally expanded and 150 tumor-infiltrating TCR groups, which revealed distinct patterns of TRBV usage, HLA association, and TCR diversity. In gastric adenocarcinoma (STAD), patients with tumor-infiltrating TCRs (Patients-TI) had significantly worse prognosis than other patients (Patients-nonTI). Patients-TI had richer CD8+ T cells in the immune microenvironment, and their gene expression features were positively correlated with immunotherapy response. We also found that tumor-infiltrating TCR groups were associated with four distinct tumor subtypes, 26 common gene mutations, and 39 gene expression signatures. We discovered that tumor-infiltrating TCRs had cross-reactivity with viral antigens, indicating a possible link between viral infections and tumor immunity.
Conclusion: By applying GLIPH2 to TCR sequences from digestive tract tumors, we uncovered novel insights into the tumor immune landscape and identified potential candidates for shared TCRs and neoantigens.
Keywords: Cross-reactivity; Digestive tract cancer; GLIPH2; Immunotherapy; TCR; Tumor antigen; Tumor immune microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Authors declare that they have no conflict of interests.
Figures




References
-
- Atkins D, Breuckmann A, Schmahl GE et al (2004) MHC class I antigen processing pathway defects, ras mutations and disease stage in colorectal carcinoma. Int J Cancer 109:265–273 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

