Coactivator condensation drives cardiovascular cell lineage specification
- PMID: 38489358
- PMCID: PMC10942106
- DOI: 10.1126/sciadv.adk7160
Coactivator condensation drives cardiovascular cell lineage specification
Abstract
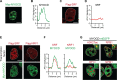
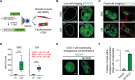
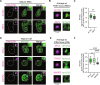
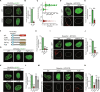
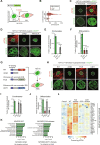
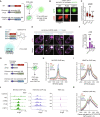
During development, cells make switch-like decisions to activate new gene programs specifying cell lineage. The mechanisms underlying these decisive choices remain unclear. Here, we show that the cardiovascular transcriptional coactivator myocardin (MYOCD) activates cell identity genes by concentration-dependent and switch-like formation of transcriptional condensates. MYOCD forms such condensates and activates cell identity genes at critical concentration thresholds achieved during smooth muscle cell and cardiomyocyte differentiation. The carboxyl-terminal disordered region of MYOCD is necessary and sufficient for condensate formation. Disrupting this region's ability to form condensates disrupts gene activation and smooth muscle cell reprogramming. Rescuing condensate formation by replacing this region with disordered regions from functionally unrelated proteins rescues gene activation and smooth muscle cell reprogramming. Our findings demonstrate that MYOCD condensate formation is required for gene activation during cardiovascular differentiation. We propose that the formation of transcriptional condensates at critical concentrations of cell type-specific regulators provides a molecular switch underlying the activation of key cell identity genes during development.
Figures


References
-
- Spiegelman B. M., Heinrich R., Biological control through regulated transcriptional coactivators. Cell 119, 157–167 (2004). - PubMed
-
- Graf T., Enver T., Forcing cells to change lineages. Nature 462, 587–594 (2009). - PubMed
-
- Palii C. G., Cheng Q., Gillespie M. A., Shannon P., Mazurczyk M., Napolitani G., Price N. D., Ranish J. A., Morrissey E., Higgs D. R., Brand M., Single-cell proteomics reveal that quantitative changes in co-expressed lineage-specific transcription factors determine cell fate. Cell Stem Cell 24, 812–820.e5 (2019). - PMC - PubMed
-
- Faure L., Wang Y., Kastriti M. E., Fontanet P., Cheung K. K. Y., Petitpre C., Wu H., Sun L. L., Runge K., Croci L., Landy M. A., Lai H. C., Consalez G. G., de Chevigny A., Lallemend F., Adameyko I., Hadjab S., Single cell RNA sequencing identifies early diversity of sensory neurons forming via bi-potential intermediates. Nat. Commun. 11, 4175 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

