Mono-UFMylation promotes misfolding-associated secretion of α-synuclein
- PMID: 38489364
- PMCID: PMC10942102
- DOI: 10.1126/sciadv.adk2542
Mono-UFMylation promotes misfolding-associated secretion of α-synuclein
Abstract
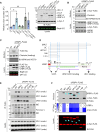
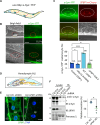
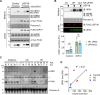
Stressed cells secret misfolded proteins lacking signaling sequence via an unconventional protein secretion (UcPS) pathway, but how misfolded proteins are targeted selectively in UcPS is unclear. Here, we report that misfolded UcPS clients are subject to modification by a ubiquitin-like protein named ubiquitin-fold modifier 1 (UFM1). Using α-synuclein (α-Syn) as a UcPS model, we show that mutating the UFMylation sites in α-Syn or genetic inhibition of the UFMylation system mitigates α-Syn secretion, whereas overexpression of UFBP1, a component of the endoplasmic reticulum-associated UFMylation ligase complex, augments α-Syn secretion in mammalian cells and in model organisms. UFM1 itself is cosecreted with α-Syn, and the serum UFM1 level correlates with that of α-Syn. Because UFM1 can be directly recognized by ubiquitin specific peptidase 19 (USP19), a previously established UcPS stimulator known to associate with several chaperoning activities, UFMylation might facilitate substrate engagement by USP19, allowing stringent and regulated selection of misfolded proteins for secretion and proteotoxic stress alleviation.
Figures



References
-
- Rabouille C., Pathways of unconventional protein secretion. Trends Cell Biol. 27, 230–240 (2017). - PubMed
-
- Nickel W., Rabouille C., Unconventional protein secretion: Diversity and consensus. Semin. Cell Dev. Biol. 83, 1–2 (2018). - PubMed
-
- Steringer J. P., Lange S., Čujova S., Šachl R., Poojari C., Lolicato F., Beutel O., Müller H. M., Unger S., Coskun Ü., Honigmann A., Vattulainen I., Hof M., Freund C., Nickel W., Key steps in unconventional secretion of fibroblast growth factor 2 reconstituted with purified components. eLife 6, e28985 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

