Increased genomic instability and reshaping of tissue microenvironment underlie oncogenic properties of Arid1a mutations
- PMID: 38489371
- PMCID: PMC10942108
- DOI: 10.1126/sciadv.adh4435
Increased genomic instability and reshaping of tissue microenvironment underlie oncogenic properties of Arid1a mutations
Abstract
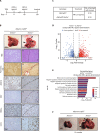
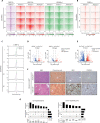
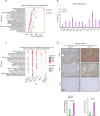
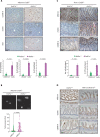
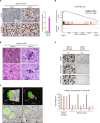
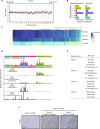
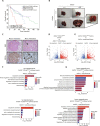
Oncogenic mutations accumulating in many chromatin-associated proteins have been identified in different tumor types. With a mutation rate from 10 to 57%, ARID1A has been widely considered a tumor suppressor gene. However, whether this role is mainly due to its transcriptional-related activities or its ability to preserve genome integrity is still a matter of intense debate. Here, we show that ARID1A is largely dispensable for preserving enhancer-dependent transcriptional regulation, being ARID1B sufficient and required to compensate for ARID1A loss. We provide in vivo evidence that ARID1A is mainly required to preserve genomic integrity in adult tissues. ARID1A loss primarily results in DNA damage accumulation, interferon type I response activation, and chronic inflammation leading to tumor formation. Our data suggest that in healthy tissues, the increased genomic instability that follows ARID1A mutations and the selective pressure imposed by the microenvironment might result in the emergence of aggressive, possibly immune-resistant, tumors.
Figures

References
-
- Centore R. C., Sandoval G. J., Soares L. M. M., Kadoch C., Chan H. M., Mammalian SWI/SNF chromatin remodeling complexes: Emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020). - PubMed
-
- Cenik B. K., Shilatifard A., COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58 (2021). - PubMed
-
- Xu G., Chhangawala S., Cocco E., Razavi P., Cai Y., Otto J. E., Ferrando L., Selenica P., Ladewig E., Chan C., da Cruz Paula A., Witkin M., Cheng Y., Park J., Serna-Tamayo C., Zhao H. Y., Wu F., Sallaku M., Qu X., Zhao A., Collings C. K., D’Avino A. R., Jhaveri K., Koche R., Levine R. L., Reis-Filho J. S., Kadoch C., Scaltriti M., Leslie C. S., Baselga J., Toska E., ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet. 52, 198–207 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

