Type-I-interferon-responsive microglia shape cortical development and behavior
- PMID: 38490196
- PMCID: PMC11015974
- DOI: 10.1016/j.cell.2024.02.020
Type-I-interferon-responsive microglia shape cortical development and behavior
Abstract
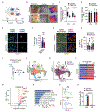
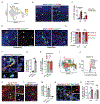
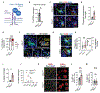
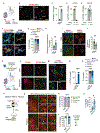
Microglia are brain-resident macrophages that shape neural circuit development and are implicated in neurodevelopmental diseases. Multiple microglial transcriptional states have been defined, but their functional significance is unclear. Here, we identify a type I interferon (IFN-I)-responsive microglial state in the developing somatosensory cortex (postnatal day 5) that is actively engulfing whole neurons. This population expands during cortical remodeling induced by partial whisker deprivation. Global or microglial-specific loss of the IFN-I receptor resulted in microglia with phagolysosomal dysfunction and an accumulation of neurons with nuclear DNA damage. IFN-I gain of function increased neuronal engulfment by microglia in both mouse and zebrafish and restricted the accumulation of DNA-damaged neurons. Finally, IFN-I deficiency resulted in excess cortical excitatory neurons and tactile hypersensitivity. These data define a role for neuron-engulfing microglia during a critical window of brain development and reveal homeostatic functions of a canonical antiviral signaling pathway in the brain.
Keywords: cortical development; microglia; neuroimmunity; phagocytosis; somatosensory cortex; tactile hypersensitivity; type I interferon.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
Type I interferon responsive microglia shape cortical development and behavior.bioRxiv [Preprint]. 2023 Mar 20:2021.04.29.441889. doi: 10.1101/2021.04.29.441889. bioRxiv. 2023. Update in: Cell. 2024 Apr 11;187(8):1936-1954.e24. doi: 10.1016/j.cell.2024.02.020. PMID: 35233577 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

