Multimodal stimulation screens reveal unique and shared genes limiting T cell fitness
- PMID: 38490212
- PMCID: PMC11003465
- DOI: 10.1016/j.ccell.2024.02.016
Multimodal stimulation screens reveal unique and shared genes limiting T cell fitness
Abstract
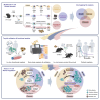
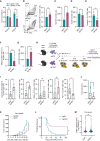
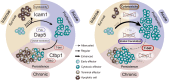
Genes limiting T cell antitumor activity may serve as therapeutic targets. It has not been systematically studied whether there are regulators that uniquely or broadly contribute to T cell fitness. We perform genome-scale CRISPR-Cas9 knockout screens in primary CD8 T cells to uncover genes negatively impacting fitness upon three modes of stimulation: (1) intense, triggering activation-induced cell death (AICD); (2) acute, triggering expansion; (3) chronic, causing dysfunction. Besides established regulators, we uncover genes controlling T cell fitness either specifically or commonly upon differential stimulation. Dap5 ablation, ranking highly in all three screens, increases translation while enhancing tumor killing. Loss of Icam1-mediated homotypic T cell clustering amplifies cell expansion and effector functions after both acute and intense stimulation. Lastly, Ctbp1 inactivation induces functional T cell persistence exclusively upon chronic stimulation. Our results functionally annotate fitness regulators based on their unique or shared contribution to traits limiting T cell antitumor activity.
Keywords: CRISPR-Cas9 screen; Ctbp1; Dap5; Icam1; T cells; activation-induced cell death; cancer immunotherapy; dysfunction; effector function; exhaustion.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.L., P.L., A.A., G.A., M.A.L., D.W.V., and D.S.P. are named as inventors on a patent filed by the Netherlands Cancer Institute and Oncode on the findings described in this manuscript. D.S.P. and M.A.L. are co-founders, shareholders, and advisors of Immagene.
Figures




References
-
- Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., Chow L.Q., Vokes E.E., Felip E., Holgado E., et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015;373:1627–1639. doi: 10.1056/NEJMOA1507643/SUPPL_FILE/NEJMOA1507643_APPENDIX.PDF. - DOI - PMC - PubMed
-
- Van Den Berg J.H., Heemskerk B., Van Rooij N., Gomez-Eerland R., Michels S., Van Zon M., De Boer R., Bakker N.A.M., Jorritsma-Smit A., Van Buuren M.M., et al. Tumor infiltrating lymphocytes (TIL) therapy in metastatic melanoma: boosting of neoantigen-specific T cell reactivity and long-term follow-up. J. Immunother. Cancer. 2020;8:e000848. doi: 10.1136/JITC-2020-000848. - DOI - PMC - PubMed
-
- Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F., et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMOA1407222/SUPPL_FILE/NEJMOA1407222_DISCLOSURES.PDF. - DOI - PMC - PubMed
-
- Dudley M.E., Wunderlich J.R., Robbins P.F., Yang J.C., Hwu P., Schwartzentruber D.J., Topalian S.L., Sherry R., Restifo N.P., Hubicki A.M., et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/SCIENCE.1076514/SUPPL_FILE/DUDLEY.SOM.PDF. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

