Translation efficiency driven by CNOT3 subunit of the CCR4-NOT complex promotes leukemogenesis
- PMID: 38491013
- PMCID: PMC10943099
- DOI: 10.1038/s41467-024-46665-2
Translation efficiency driven by CNOT3 subunit of the CCR4-NOT complex promotes leukemogenesis
Erratum in
-
Publisher Correction: Translation efficiency driven by CNOT3 subunit of the CCR4-NOT complex promotes leukemogenesis.Nat Commun. 2024 Oct 7;15(1):8674. doi: 10.1038/s41467-024-53071-1. Nat Commun. 2024. PMID: 39375334 Free PMC article. No abstract available.
Abstract
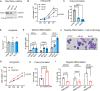
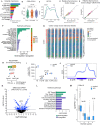
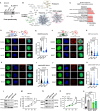
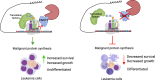
Protein synthesis is frequently deregulated during tumorigenesis. However, the precise contexts of selective translational control and the regulators of such mechanisms in cancer is poorly understood. Here, we uncovered CNOT3, a subunit of the CCR4-NOT complex, as an essential modulator of translation in myeloid leukemia. Elevated CNOT3 expression correlates with unfavorable outcomes in patients with acute myeloid leukemia (AML). CNOT3 depletion induces differentiation and apoptosis and delayed leukemogenesis. Transcriptomic and proteomic profiling uncovers c-MYC as a critical downstream target which is translationally regulated by CNOT3. Global analysis of mRNA features demonstrates that CNOT3 selectively influences expression of target genes in a codon usage dependent manner. Furthermore, CNOT3 associates with the protein network largely consisting of ribosomal proteins and translation elongation factors in leukemia cells. Overall, our work elicits the direct requirement for translation efficiency in tumorigenesis and propose targeting the post-transcriptional circuitry via CNOT3 as a therapeutic vulnerability in AML.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

