Notoginsenoside R1 promotes Lgr5+ stem cell and epithelium renovation in colitis mice via activating Wnt/β-Catenin signaling
- PMID: 38491161
- PMCID: PMC11192909
- DOI: 10.1038/s41401-024-01250-7
Notoginsenoside R1 promotes Lgr5+ stem cell and epithelium renovation in colitis mice via activating Wnt/β-Catenin signaling
Abstract
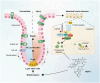
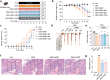
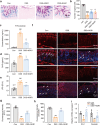
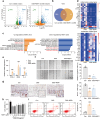
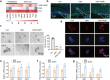
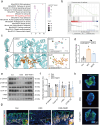
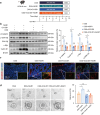
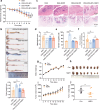
Inflammatory bowel disease (IBD) is characterized by persistent damage to the intestinal barrier and excessive inflammation, leading to increased intestinal permeability. Current treatments of IBD primarily address inflammation, neglecting epithelial repair. Our previous study has reported the therapeutic potential of notoginsenoside R1 (NGR1), a characteristic saponin from the root of Panax notoginseng, in alleviating acute colitis by reducing mucosal inflammation. In this study we investigated the reparative effects of NGR1 on mucosal barrier damage after the acute injury stage of DSS exposure. DSS-induced colitis mice were orally treated with NGR1 (25, 50, 125 mg·kg-1·d-1) for 10 days. Body weight and rectal bleeding were daily monitored throughout the experiment, then mice were euthanized, and the colon was collected for analysis. We showed that NGR1 administration dose-dependently ameliorated mucosal inflammation and enhanced epithelial repair evidenced by increased tight junction proteins, mucus production and reduced permeability in colitis mice. We then performed transcriptomic analysis on rectal tissue using RNA-sequencing, and found NGR1 administration stimulated the proliferation of intestinal crypt cells and facilitated the repair of epithelial injury; NGR1 upregulated ISC marker Lgr5, the genes for differentiation of intestinal stem cells (ISCs), as well as BrdU incorporation in crypts of colitis mice. In NCM460 human intestinal epithelial cells in vitro, treatment with NGR1 (100 μM) promoted wound healing and reduced cell apoptosis. NGR1 (100 μM) also increased Lgr5+ cells and budding rates in a 3D intestinal organoid model. We demonstrated that NGR1 promoted ISC proliferation and differentiation through activation of the Wnt signaling pathway. Co-treatment with Wnt inhibitor ICG-001 partially counteracted the effects of NGR1 on crypt Lgr5+ ISCs, organoid budding rates, and overall mice colitis improvement. These results suggest that NGR1 alleviates DSS-induced colitis in mice by promoting the regeneration of Lgr5+ stem cells and intestinal reconstruction, at least partially via activation of the Wnt/β-Catenin signaling pathway. Schematic diagram of the mechanism of NGR1 in alleviating colitis. DSS caused widespread mucosal inflammation epithelial injury. This was manifested by the decreased expression of tight junction proteins, reduced mucus production in goblet cells, and increased intestinal permeability in colitis mice. Additionally, Lgr5+ ISCs were in obviously deficiency in colitis mice, with aberrant down-regulation of the Wnt/β-Catenin signaling. However, NGR1 amplified the expression of the ISC marker Lgr5, elevated the expression of genes associated with ISC differentiation, enhanced the incorporation of BrdU in the crypt and promoted epithelial restoration to alleviate DSS-induced colitis in mice, at least partially, by activating the Wnt/β-Catenin signaling pathway.
Keywords: ICG-001; Wnt/β-Catenin pathway; inflammatory bowel disease; intestinal stem cells; notoginsenoside R1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

