Appropriate glycemic management protects the germline but not the uterine environment in hyperglycemia
- PMID: 38491313
- PMCID: PMC11014859
- DOI: 10.1038/s44319-024-00097-7
Appropriate glycemic management protects the germline but not the uterine environment in hyperglycemia
Abstract
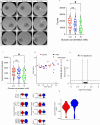
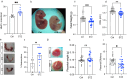
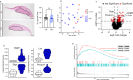
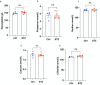
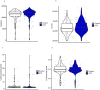
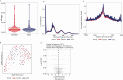
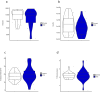
Emerging evidence indicates that parental diseases can impact the health of subsequent generations through epigenetic inheritance. Recently, it was shown that maternal diabetes alters the metaphase II oocyte transcriptome, causing metabolic dysfunction in offspring. However, type 1 diabetes (T1D) mouse models frequently utilized in previous studies may be subject to several confounding factors due to severe hyperglycemia. This limits clinical translatability given improvements in glycemic control for T1D subjects. Here, we optimize a T1D mouse model to investigate the effects of appropriately managed maternal glycemic levels on oocytes and intrauterine development. We show that diabetic mice with appropriate glycemic control exhibit better long-term health, including maintenance of the oocyte transcriptome and chromatin accessibility. We further show that human oocytes undergoing in vitro maturation challenged with mildly increased levels of glucose, reflecting appropriate glycemic management, also retain their transcriptome. However, fetal growth and placental function are affected in mice despite appropriate glycemic control, suggesting the uterine environment rather than the germline as a pathological factor in developmental programming in appropriately managed diabetes.
Keywords: Developmental Programming; Diabetes; Fetal Development; Hypoxia; Placenta.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Beck RW, Riddlesworth T, Ruedy K, Ahmann A, Bergenstal R, Haller S, Kollman C, Kruger D, McGill JB, Polonsky W, et al. Effect of continuous glucose monitoring on glycemic control in adults with type 1 diabetes using insulin injections: the DIAMOND randomized clinical trial. JAMA. 2017;317:371–378. doi: 10.1001/jama.2016.19975. - DOI - PubMed
-
- Bergenstal RM, Nimri R, Beck RW, Criego A, Laffel L, Schatz D, Battelino T, Danne T, Weinzimer SA, Sibayan J, et al. A comparison of two hybrid closed-loop systems in adolescents and young adults with type 1 diabetes (FLAIR): a multicentre, randomised, crossover trial. Lancet Lond Engl. 2021;397:208–219. doi: 10.1016/S0140-6736(20)32514-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

