Pitting the olive seed microbiome
- PMID: 38491515
- PMCID: PMC10943921
- DOI: 10.1186/s40793-024-00560-x
Pitting the olive seed microbiome
Abstract
Background: The complex and co-evolved interplay between plants and their microbiota is crucial for the health and fitness of the plant holobiont. However, the microbiota of the seeds is still relatively unexplored and no studies have been conducted with olive trees so far. In this study, we aimed to characterize the bacterial, fungal and archaeal communities present in seeds of ten olive genotypes growing in the same orchard through amplicon sequencing to test whether the olive genotype is a major driver in shaping the seed microbial community, and to identify the origin of the latter. Therefore, we have developed a methodology for obtaining samples from the olive seed's endosphere under sterile conditions.
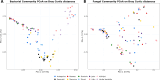
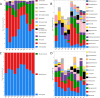
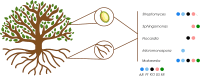
Results: A diverse microbiota was uncovered in olive seeds, the plant genotype being an important factor influencing the structure and composition of the microbial communities. The most abundant bacterial phylum was Actinobacteria, accounting for an average relative abundance of 41%. At genus level, Streptomyces stood out because of its potential influence on community structure. Within the fungal community, Basidiomycota and Ascomycota were the most abundant phyla, including the genera Malassezia, Cladosporium, and Mycosphaerella. The shared microbiome was composed of four bacterial (Stenotrophomonas, Streptomyces, Promicromonospora and Acidipropionibacterium) and three fungal (Malassezia, Cladosporium and Mycosphaerella) genera. Furthermore, a comparison between findings obtained here and earlier results from the root endosphere of the same trees indicated that genera such as Streptomyces and Malassezia were present in both olive compartments.
Conclusions: This study provides the first insights into the composition of the olive seed microbiota. The highly abundant fungal genus Malassezia and the bacterial genus Streptomyces reflect a unique signature of the olive seed microbiota. The genotype clearly shaped the composition of the seed's microbial community, although a shared microbiome was found. We identified genera that may translocate from the roots to the seeds, as they were present in both organs of the same trees. These findings set the stage for future research into potential vertical transmission of olive endophytes and the role of specific microbial taxa in seed germination, development, and seedling survival.
Keywords: Cladosporium; Malassezia; Olea europaea; Streptomyces; Olive genotypes; Vertical transmission.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bergna A, Cernava T, Rändler M, Grosch R, Zachow C, Berg G. Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J. 2018;2:183–93. doi: 10.1094/PBIOMES-06-18-0029-R. - DOI
LinkOut - more resources
Full Text Sources
