Testing the feasibility of targeting a conserved region on the S2 domain of the SARS-CoV-2 spike protein
- PMID: 38491772
- PMCID: PMC11052916
- DOI: 10.1016/j.bpj.2024.03.018
Testing the feasibility of targeting a conserved region on the S2 domain of the SARS-CoV-2 spike protein
Abstract
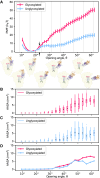
The efficacy of vaccines against the SARS-CoV-2 virus significantly declines with the emergence of mutant strains, prompting investigation into the feasibility of targeting highly conserved but often cryptic regions on the S2 domain of spike protein. Using tools from molecular dynamics, we find that exposure of a conserved S2 epitope located in the central helices below the receptor binding domains would require large-scale motion beyond receptor binding domain up-down motion, but, along the reaction coordinates we explored, it is unlikely to be exposed by such large-scale dynamic fluctuations of the S1 domain without any external facilitating factors, despite some previous computational evidence suggesting transient exposure of this region. Furthermore, glycans, particularly those on N165 and N234, hinder S2-exposing opening dynamics, and thus stabilize spike in addition to immunologically shielding the protein surface. Although the S2 epitope region examined here is central to large-scale conformational changes during viral entry, free energy landscape analysis obtained using the path coordinate formalism reveals no inherent "loaded spring" effect, suggesting that a vaccine immunogen would tend to present the epitope in a prefusion-like conformation and may be effective in neutralization. These findings contribute to a deeper understanding of the dynamic origins of the function of the spike protein, as well as further characterizing the feasibility of the S2 epitope as a therapeutic target.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests P.G., S.C.C.H., and S.S.P. are co-inventors on US patent application no. PCT/CA2024/050214 entitled “Evolutionarily Conserved Epitopes in Beta-Coronaviruses and Vaccines and Antibodies Thereto.”
Figures
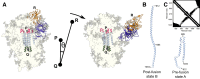

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

