A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides
- PMID: 38491946
- PMCID: PMC10979747
- DOI: 10.1021/jacs.3c12037
A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides
Abstract
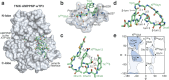
Thiopeptides make up a group of structurally complex peptidic natural products holding promise in bioengineering applications. The previously established thiopeptide/mRNA display platform enables de novo discovery of natural product-like thiopeptides with designed bioactivities. However, in contrast to natural thiopeptides, the discovered structures are composed predominantly of proteinogenic amino acids, which results in low metabolic stability in many cases. Here, we redevelop the platform and demonstrate that the utilization of compact reprogrammed genetic codes in mRNA display libraries can lead to the discovery of thiopeptides predominantly composed of nonproteinogenic structural elements. We demonstrate the feasibility of our designs by conducting affinity selections against Traf2- and NCK-interacting kinase (TNIK). The experiment identified a series of thiopeptides with high affinity to the target protein (the best KD = 2.1 nM) and kinase inhibitory activity (the best IC50 = 0.15 μM). The discovered compounds, which bore as many as 15 nonproteinogenic amino acids in an 18-residue macrocycle, demonstrated high metabolic stability in human serum with a half-life of up to 99 h. An X-ray cocrystal structure of TNIK in complex with a discovered thiopeptide revealed how nonproteinogenic building blocks facilitate the target engagement and orchestrate the folding of the thiopeptide into a noncanonical conformation. Altogether, the established platform takes a step toward the discovery of thiopeptides with high metabolic stability for early drug discovery applications.
Conflict of interest statement
The authors declare the following competing financial interest(s): A.V., Y.Z., Y.G. and H.S. are listed as co-inventors on the patents pertaining to lactazole engineering. Other authors declare no competing interests.
Figures





Similar articles
-
De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.J Am Chem Soc. 2022 Nov 9;144(44):20332-20341. doi: 10.1021/jacs.2c07937. Epub 2022 Oct 25. J Am Chem Soc. 2022. PMID: 36282922 Free PMC article.
-
Recombinant thiopeptides containing noncanonical amino acids.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3615-20. doi: 10.1073/pnas.1602733113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976568 Free PMC article.
-
Deep Learning-Driven Library Design for the De Novo Discovery of Bioactive Thiopeptides.ACS Cent Sci. 2023 Nov 7;9(11):2150-2160. doi: 10.1021/acscentsci.3c00957. eCollection 2023 Nov 22. ACS Cent Sci. 2023. PMID: 38033794 Free PMC article.
-
Elucidating and engineering thiopeptide biosynthesis.World J Microbiol Biotechnol. 2017 Jun;33(6):119. doi: 10.1007/s11274-017-2283-9. Epub 2017 May 11. World J Microbiol Biotechnol. 2017. PMID: 28497389 Review.
-
Introduction to Thiopeptides: Biological Activity, Biosynthesis, and Strategies for Functional Reprogramming.Cell Chem Biol. 2020 Aug 20;27(8):1032-1051. doi: 10.1016/j.chembiol.2020.07.003. Epub 2020 Jul 21. Cell Chem Biol. 2020. PMID: 32698017 Review.
Cited by
-
Ligand-Enabled Selective Coupling of MIDA Boronates to Dehydroalanine-Containing Peptides and Proteins.J Am Chem Soc. 2025 Mar 5;147(9):7533-7544. doi: 10.1021/jacs.4c16525. Epub 2025 Feb 21. J Am Chem Soc. 2025. PMID: 39984172
-
Cell-free synthetic biology for natural product biosynthesis and discovery.Chem Soc Rev. 2025 May 6;54(9):4314-4352. doi: 10.1039/d4cs01198h. Chem Soc Rev. 2025. PMID: 40104998 Free PMC article. Review.
-
Expanded ribosomal synthesis of non-standard cyclic backbones in vitro.Nat Commun. 2025 May 28;16(1):4957. doi: 10.1038/s41467-025-60126-4. Nat Commun. 2025. PMID: 40436849 Free PMC article.
References
-
- Montalbán-López M.; Scott T. A.; Ramesh S.; Rahman I. R.; van Heel A. J.; Viel J. H.; Bandarian V.; Dittmann E.; Genilloud O.; Goto Y.; Grande Burgos M. J.; Hill C.; Kim S.; Koehnke J.; Latham J. A.; Link A. J.; Martínez B.; Nair S. K.; Nicolet Y.; Rebuffat S.; Sahl H.-G.; Sareen D.; Schmidt E. W.; Schmitt L.; Severinov K.; Süssmuth R. D.; Truman A. W.; Wang H.; Weng J.-K.; van Wezel G. P.; Zhang Q.; Zhong J.; Piel J.; Mitchell D. A.; Kuipers O. P.; van der Donk W. A. New Developments in RiPP Discovery, Enzymology and Engineering. Nat. Prod. Rep. 2021, 38, 130–239. 10.1039/D0NP00027B. - DOI - PMC - PubMed
-
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.; Gruber C. W.; Fischbach M. A.; Garavelli J. S.; Ulf G.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Nair S. K.; Mitchell D. A.; Moll G. N.; Moore B. S.; Rolf M.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J. T.; Ross R. P.; Sahl H.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Tagg J. R.; Tang G.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. 10.1039/C2NP20085F. - DOI - PMC - PubMed
-
- Harms J. M.; Wilson D. N.; Schluenzen F.; Connell S. R.; Stachelhaus T.; Zaborowska Z.; Spahn C. M. T.; Fucini P. Translational Regulation via L11: Molecular Switches on the Ribosome Turned On and Off by Thiostrepton and Micrococcin. Mol. Cell 2008, 30, 26–38. 10.1016/j.molcel.2008.01.009. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

