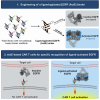
An engineering strategy to target activated EGFR with CAR T cells
- PMID: 38492569
- PMCID: PMC11045874
- DOI: 10.1016/j.crmeth.2024.100728
An engineering strategy to target activated EGFR with CAR T cells
Abstract
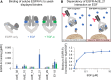
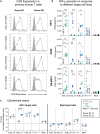
Chimeric antigen receptor (CAR) T cells have shown remarkable response rates in hematological malignancies. In contrast, CAR T cell treatment of solid tumors is associated with several challenges, in particular the expression of most tumor-associated antigens at lower levels in vital organs, resulting in on-target/off-tumor toxicities. Thus, innovative approaches to improve the tumor specificity of CAR T cells are urgently needed. Based on the observation that many human solid tumors activate epidermal growth factor receptor (EGFR) on their surface through secretion of EGFR ligands, we developed an engineering strategy for CAR-binding domains specifically directed against the ligand-activated conformation of EGFR. We show, in several experimental systems, that the generated binding domains indeed enable CAR T cells to distinguish between active and inactive EGFR. We anticipate that this engineering concept will be an important step forward to improve the tumor specificity of CAR T cells directed against EGFR-positive solid cancers.
Keywords: CAR T cells; CP: cancer biology; EGF; Nur77 reporter cell line; TGF-α; activated EGFR; antigen-binding domain; conformational specificity; protein engineering strategy; protein-protein interaction; yeast surface display technology.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.L. and M.W.T. receive funding from Miltenyi Biotec.
Figures





References
-
- Arena S., Bellosillo B., Siravegna G., Martínez A., Cañadas I., Lazzari L., Ferruz N., Russo M., Misale S., González I., et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin. Cancer Res. 2015;21:2157–2166. doi: 10.1158/1078-0432.CCR-14-2821. - DOI - PubMed
-
- Bagchi A., Haidar J.N., Eastman S.W., Vieth M., Topper M., Iacolina M.D., Walker J.M., Forest A., Shen Y., Novosiadly R.D., Ferguson K.M. Molecular Basis for Necitumumab Inhibition of EGFR Variants Associated with Acquired Cetuximab Resistance. Mol. Cancer Ther. 2018;17:521–531. doi: 10.1158/1535-7163.MCT-17-0575. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

