Structural impact of a new spike Y170W mutation detected in early emerging SARS-CoV-2 Omicron variants in France
- PMID: 38492859
- PMCID: PMC10957500
- DOI: 10.1016/j.virusres.2024.199354
Structural impact of a new spike Y170W mutation detected in early emerging SARS-CoV-2 Omicron variants in France
Abstract
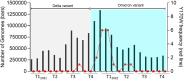
To assess the genetic characteristics of the early emerging SARS-CoV-2 Omicron variant strains, we retrospectively analyzed a collection of 150 nasopharyngeal samples taken from a series of outpatient cases tested positive by a referenced qRT-PCR assay during the reported period of Omicron variant emergence in December 2021, in northeastern region of France. Next Generation Sequencing (NGS) analysis of SARS-CoV-2 spike sequences revealed that only 3 (2 %) of these detected strains were Omicron variants, while 147 (98 %) were identified as previously described delta variants. Our phylogenetic analyzes of SARS-CoV-2 RNA genomes showed that these French early emerging Omicron variants may have originated from South Africa or India. In addition, whole viral genome sequences NGS comparison analyzes allowed us to identify an original and uncharacterized Y170W spike mutation that was weakly and transiently detected during the period of SARS-CoV-2 Omicron variant emergence in human populations. Molecular modeling and docking experiments indicated that this original mutated residue Y170W was neither directly involved in binding to the SARS-CoV-2 receptor ACE2 nor in interacting with known neutralizing antibody sites. However, this new mutation may be responsible for preventing the transition from the closed to the open Spike conformation, thus promoting the early emergence of the Omicron variant. Overall, these results underscore the epidemiological utility of a routine whole-genome viral NGS strategy that enables genotypic characterization of emerging or mutant SARS-CoV-2 variants, which could have significant implications for public health policy.
Keywords: COVID-19; Epidemiology; NGS; Omicron variant; SARS-CoV-2; Spike gene.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Characterization of the viral genome of Omicron variants of SARS-CoV-2 circulating in Tripura, a remote frontier state in Northeastern India.Mol Biol Rep. 2024 Oct 28;51(1):1100. doi: 10.1007/s11033-024-10048-z. Mol Biol Rep. 2024. PMID: 39466467
-
Molecular epidemiology of SARS-CoV-2 in Northern South Africa: wastewater surveillance from January 2021 to May 2022.Front Public Health. 2023 Dec 19;11:1309869. doi: 10.3389/fpubh.2023.1309869. eCollection 2023. Front Public Health. 2023. PMID: 38174083 Free PMC article.
-
Identification of Various Recombinants in a Patient Coinfected With the Different SARS-CoV-2 Variants.Influenza Other Respir Viruses. 2024 Jun;18(6):e13340. doi: 10.1111/irv.13340. Influenza Other Respir Viruses. 2024. PMID: 38890805 Free PMC article.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
Omicron variant in COVID-19 current pandemic: a reason for apprehension.Horm Mol Biol Clin Investig. 2022 Sep 5;44(1):89-96. doi: 10.1515/hmbci-2022-0010. eCollection 2023 Mar 1. Horm Mol Biol Clin Investig. 2022. PMID: 36064193 Review.
References
-
- Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. doi: 10.21105/joss.03773. - DOI
-
- Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. - DOI
-
- CDC Coronavirus Disease 2019 (COVID-19) [WWW Document. Cent. Dis. Control Prev. 2020 https://www.cdc.gov/coronavirus/2019-ncov/hcp/long-term-care.html URL. accessed 4.20.20.
-
- Cele, S., Jackson, L., Khoury, D.S., Khan, K., Moyo-Gwete, T., Tegally, H., San, J.E., Cromer, D., Scheepers, C., Amoako, D., Karim, F., Bernstein, M., Lustig, G., Archary, D., Smith, M., Ganga, Y., Jule, Z., Reedoy, K., Hwa, S.H., Giandhari, J., Blackburn, J.M., Gosnell, B.I., Karim, S.S.A., Hanekom, W., Ngs-Sa, Team, CK., Gottberg, A. von, Bhiman, J., Lessells, R.J., Moosa, MY.S., Davenport, M.P., Oliveira, T.de, Moore, P.L., Sigal, A., 2021. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. 10.1101/2021.12.08.21267417. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

