High-throughput sequencing reveals differences in microbial community structure and diversity in the conjunctival tissue of healthy and type 2 diabetic mice
- PMID: 38493114
- PMCID: PMC10943819
- DOI: 10.1186/s12866-024-03247-y
High-throughput sequencing reveals differences in microbial community structure and diversity in the conjunctival tissue of healthy and type 2 diabetic mice
Abstract
Background: To investigate the differences in bacterial and fungal community structure and diversity in conjunctival tissue of healthy and diabetic mice.
Methods: RNA-seq assays and high-throughput sequencing of bacterial 16 S rDNA and fungal internal transcribed spacer (ITS) gene sequences were used to identify differentially expressed host genes and fungal composition profiles in conjunctival tissues of diabetic BKS-db/db mice and BKS (control) mice. Functional enrichment analysis of differentially expressed genes and the correlation between the relative abundance of bacterial and fungal taxa in the intestinal mucosa were also performed.
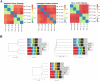
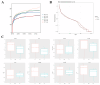
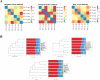
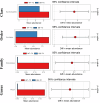
Results: Totally, 449 differential up-regulated genes and 1,006 down-regulated genes were identified in the conjunctival tissues of diabetic mice. The differentially expressed genes were mainly enriched in metabolism-related functions and pathways. A decrease in conjunctival bacterial species diversity and abundance in diabetic mice compared to control mice. In contrast, fungal species richness and diversity were not affected by diabetes. The microbial colonies were mainly associated with cellular process pathways regulating carbohydrate and lipid metabolism, as well as cell growth and death. Additionally, some interactions between bacteria and fungi at different taxonomic levels were also observed.
Conclusion: The present study revealed significant differences in the abundance and composition of bacterial and fungal communities in the conjunctival tissue of diabetic mice compared to control mice. The study also highlighted interactions between bacteria and fungi at different taxonomic levels. These findings may have implications for the diagnosis and treatment of diabetes.
Keywords: Conjunctival tissue; Diabetes mellitus; Diversity; High-throughput sequencing; Microbial community.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
High-throughput sequencing revealed differences of microbial community structure and diversity between healthy and diseased Caulerpa lentillifera.BMC Microbiol. 2019 Oct 15;19(1):225. doi: 10.1186/s12866-019-1605-5. BMC Microbiol. 2019. PMID: 31615401 Free PMC article.
-
Defining the normal core microbiome of conjunctival microbial communities.Clin Microbiol Infect. 2016 Jul;22(7):643.e7-643.e12. doi: 10.1016/j.cmi.2016.04.008. Epub 2016 Apr 19. Clin Microbiol Infect. 2016. PMID: 27102141
-
Indoor microbiota in severely moisture damaged homes and the impact of interventions.Microbiome. 2017 Oct 13;5(1):138. doi: 10.1186/s40168-017-0356-5. Microbiome. 2017. PMID: 29029638 Free PMC article.
-
Plants Play Stronger Effects on Soil Fungal than Bacterial Communities and Co-Occurrence Network Structures in a Subtropical Tree Diversity Experiment.Microbiol Spectr. 2022 Jun 29;10(3):e0013422. doi: 10.1128/spectrum.00134-22. Epub 2022 Apr 27. Microbiol Spectr. 2022. PMID: 35475656 Free PMC article.
-
Effects of Exogenous Hydrogen Sulfide on Diabetic Metabolic Disorders in db/db Mice Are Associated With Gut Bacterial and Fungal Microbiota.Front Cell Infect Microbiol. 2022 Mar 29;12:801331. doi: 10.3389/fcimb.2022.801331. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35425717 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

